Arc is on 🔥

Arc is on 🔥
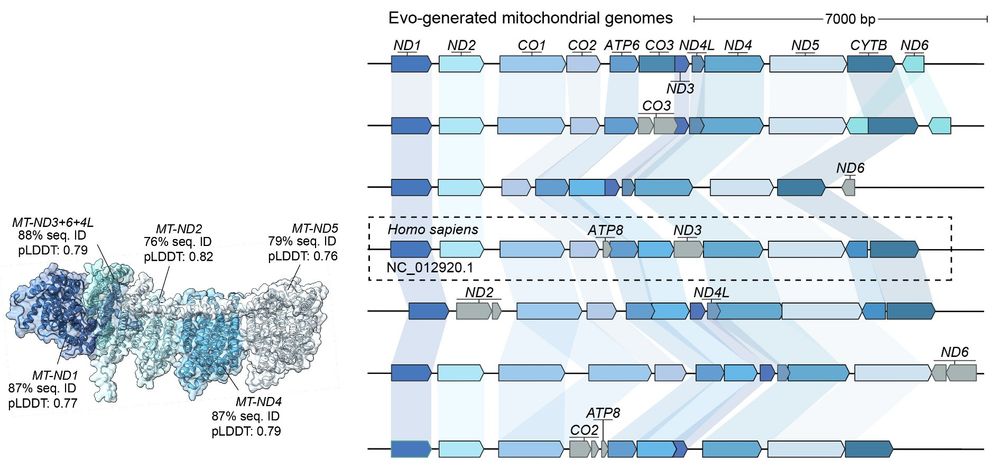
We're excited about programmable genome design at unprecedented length scales, especially when combined with AI-generated DNA sequences of high complexity (e.g. Evo 2)
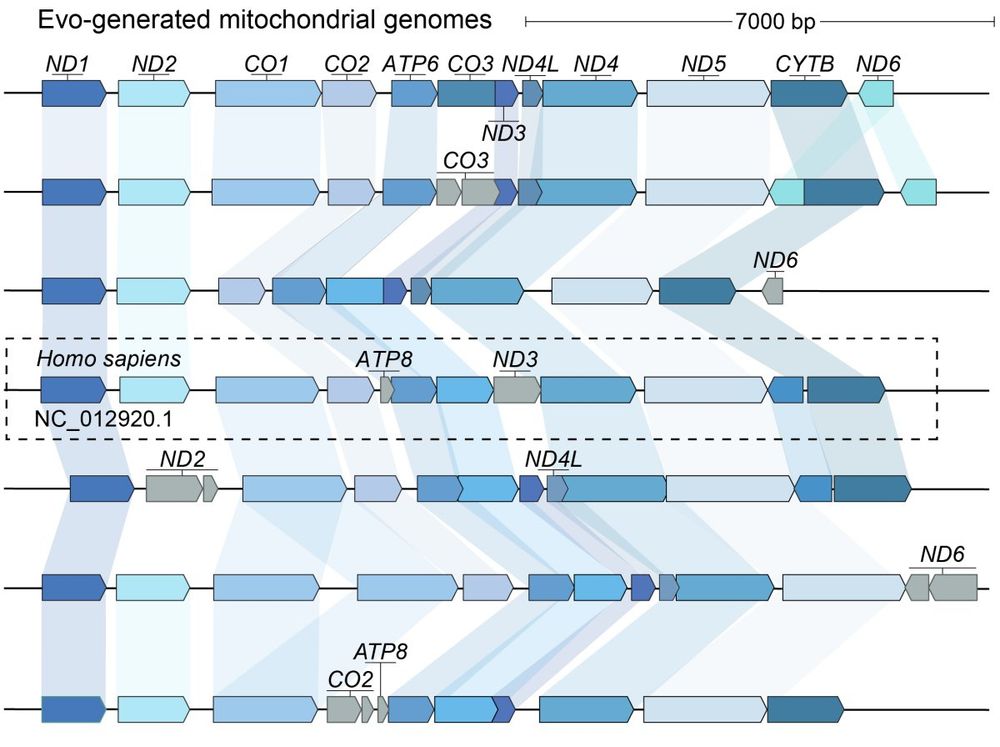
We're excited about programmable genome design at unprecedented length scales, especially when combined with AI-generated DNA sequences of high complexity (e.g. Evo 2)
We provide a therapeutic proof-of-concept with bridge-mediated excision of the BCL11A enhancer for sickle cell anemia and of expanded repeat sequences found in Friedreich's ataxia
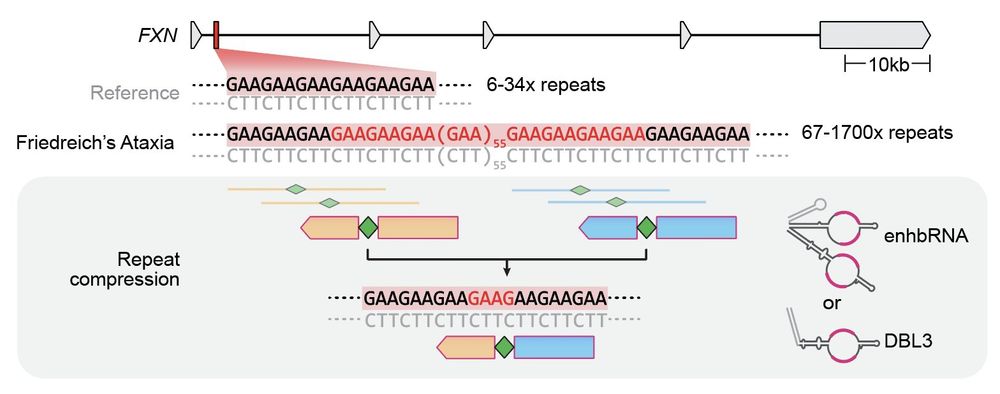
We provide a therapeutic proof-of-concept with bridge-mediated excision of the BCL11A enhancer for sickle cell anemia and of expanded repeat sequences found in Friedreich's ataxia
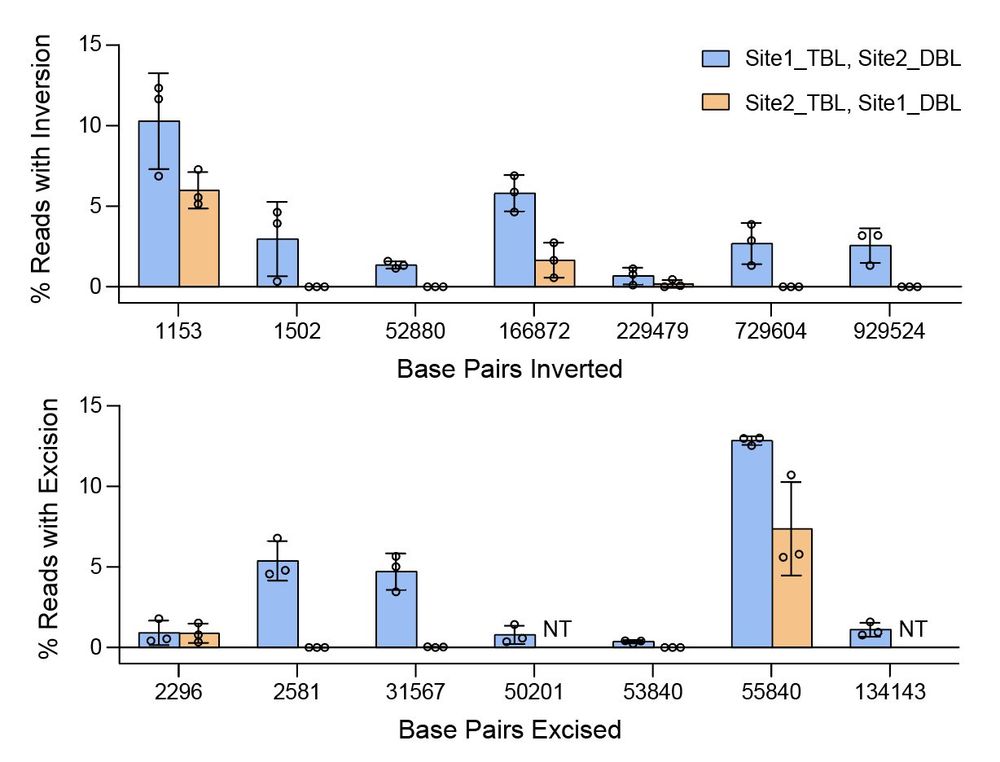
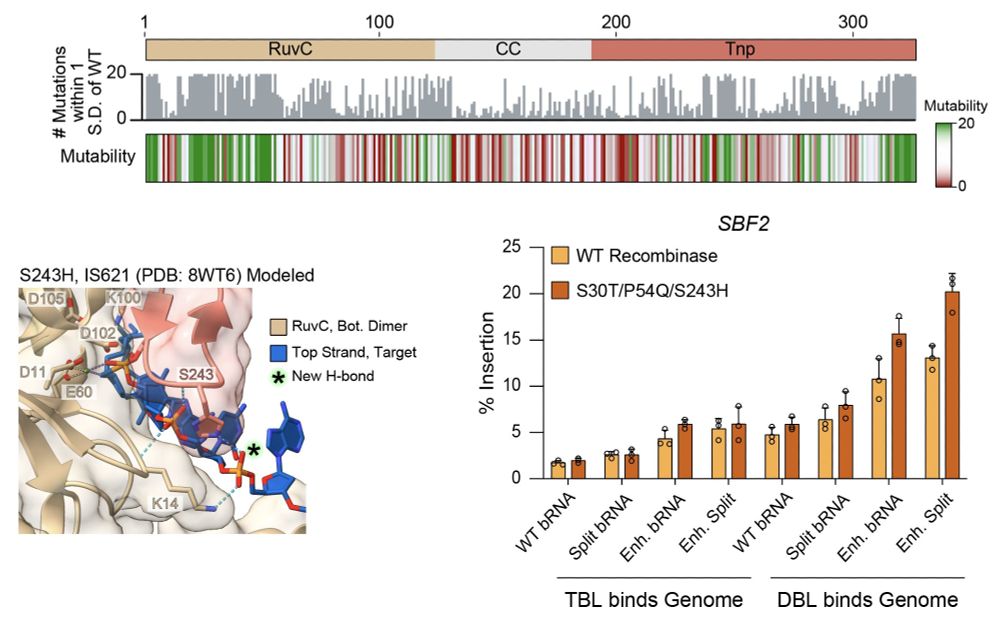
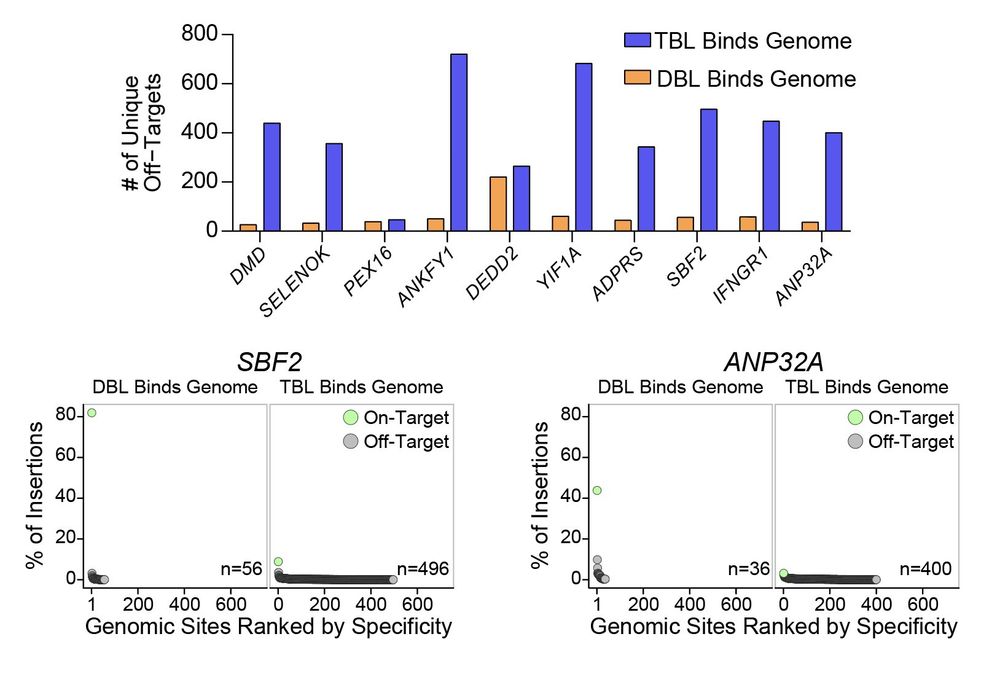
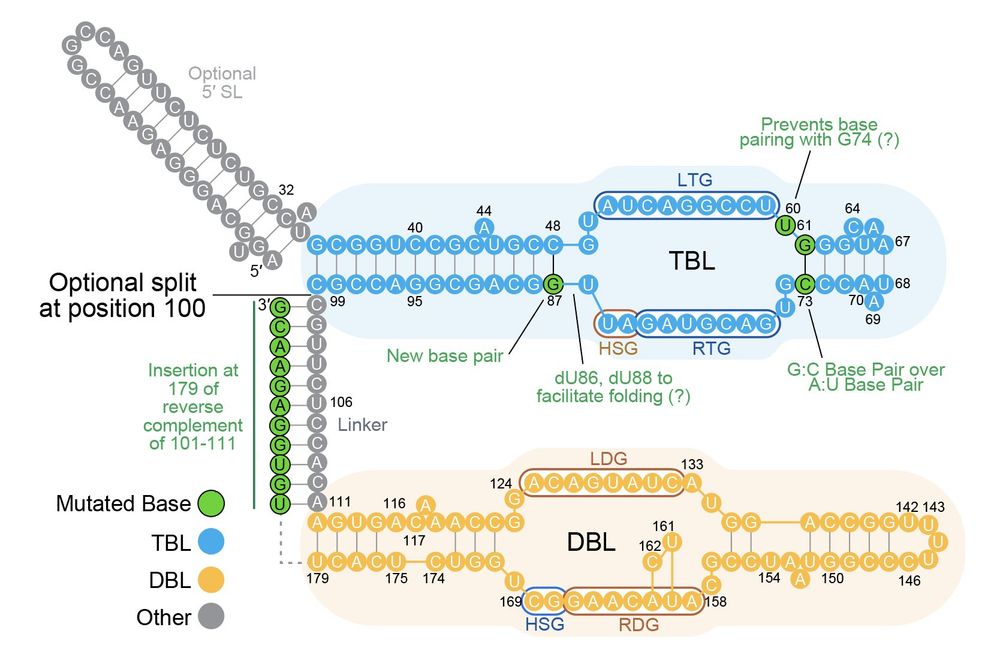
In new work @arcinstitute.org, we report the discovery and engineering of the first programmable DNA recombinases capable of megabase-scale human genome rearrangement
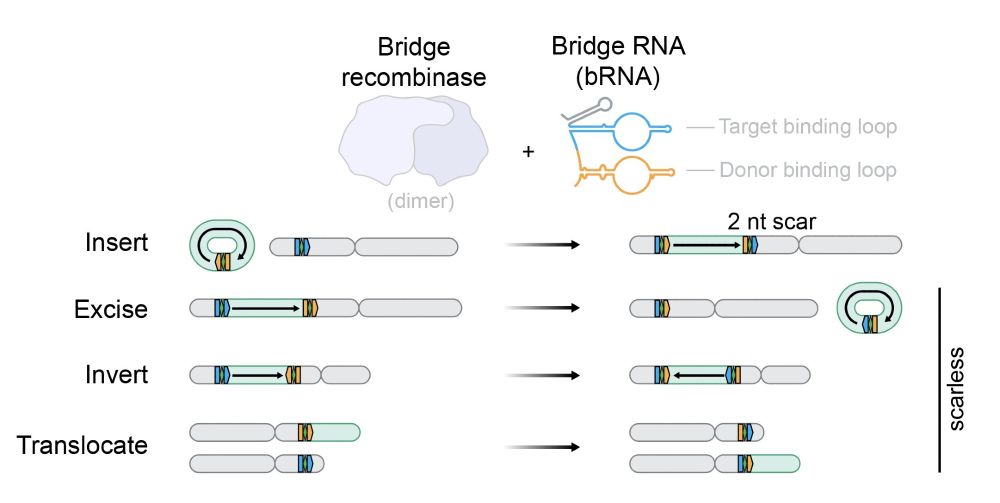
In new work @arcinstitute.org, we report the discovery and engineering of the first programmable DNA recombinases capable of megabase-scale human genome rearrangement
is hiring. Check out open Arc jobs at arcinstitute.org/jobs or just email me directly. Our research group is hiring in molecular machine learning and the interface of computational and synthetic biology

is hiring. Check out open Arc jobs at arcinstitute.org/jobs or just email me directly. Our research group is hiring in molecular machine learning and the interface of computational and synthetic biology
Intriguingly, this AI brain has features that may correspond to regulatory elements
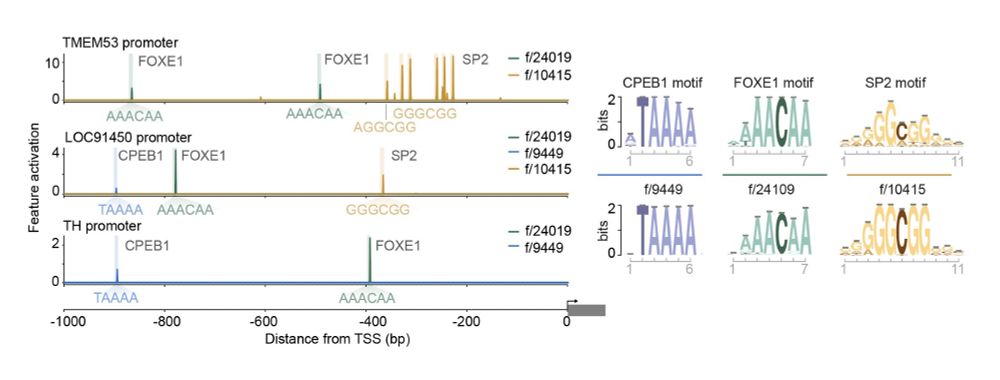
Intriguingly, this AI brain has features that may correspond to regulatory elements
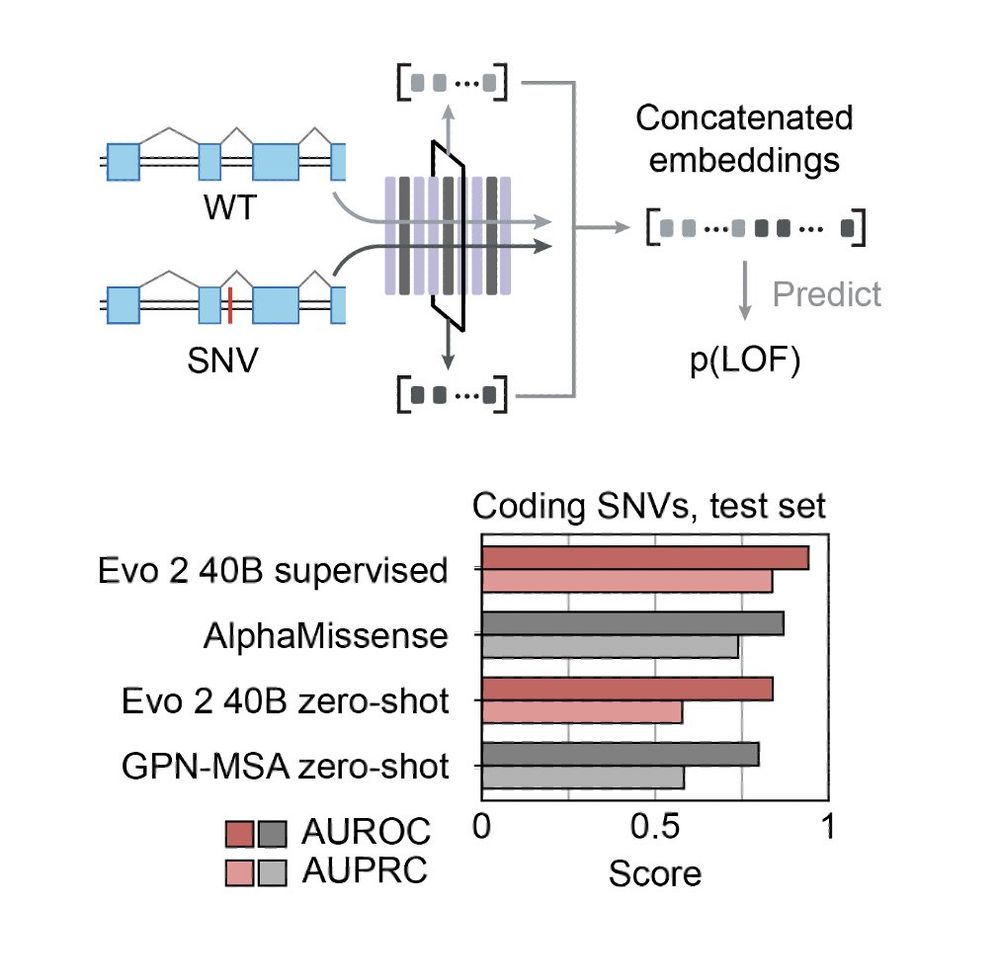
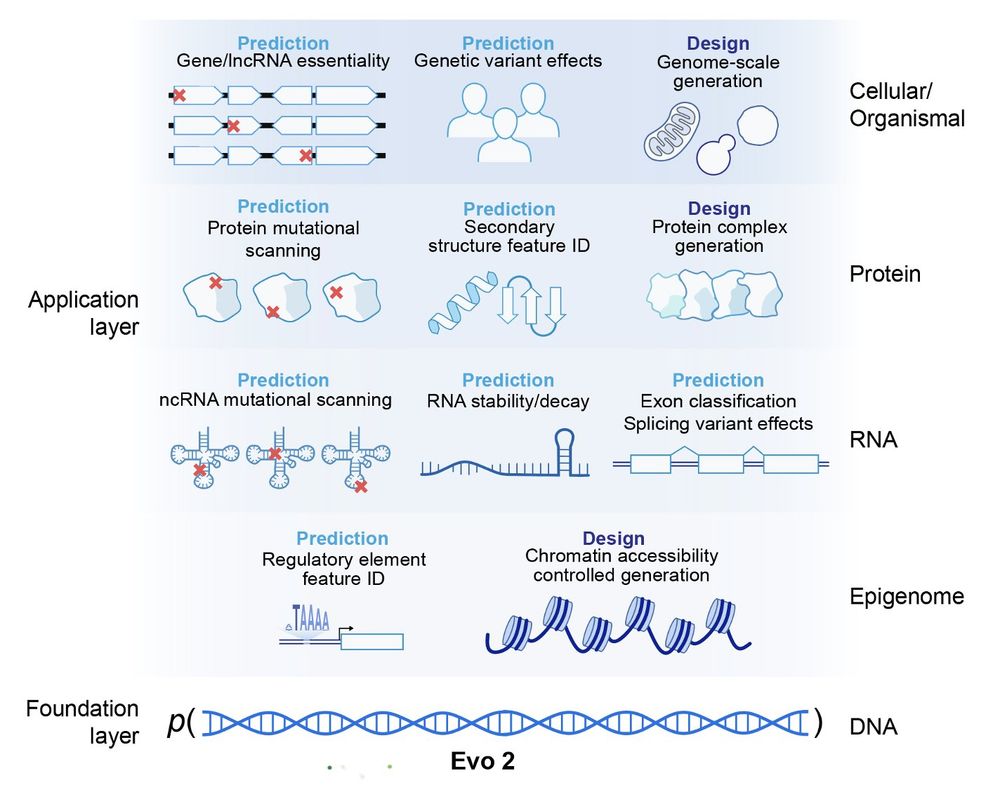
It's state of the art in doing this zero-shot for noncoding mutations
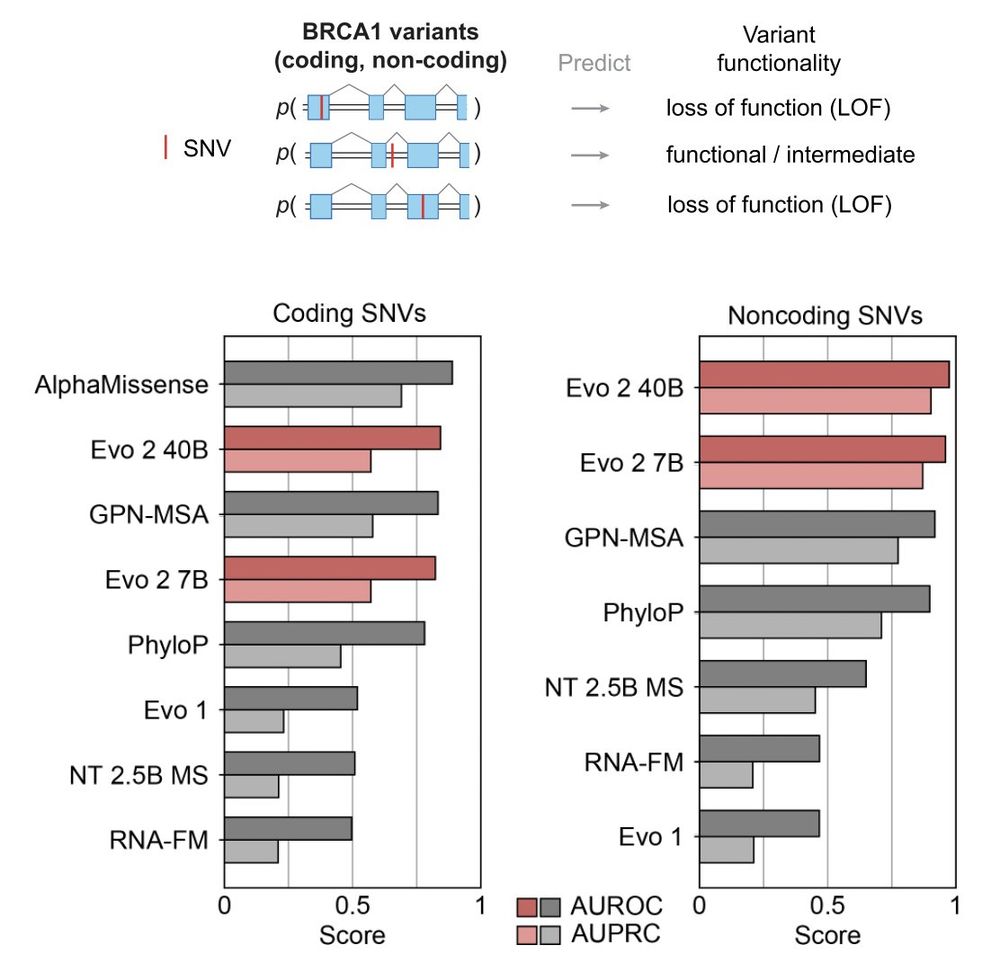
It's state of the art in doing this zero-shot for noncoding mutations
In other words, if you have a genetic mutation, Evo 2 has an opinion on whether or not it might cause disease
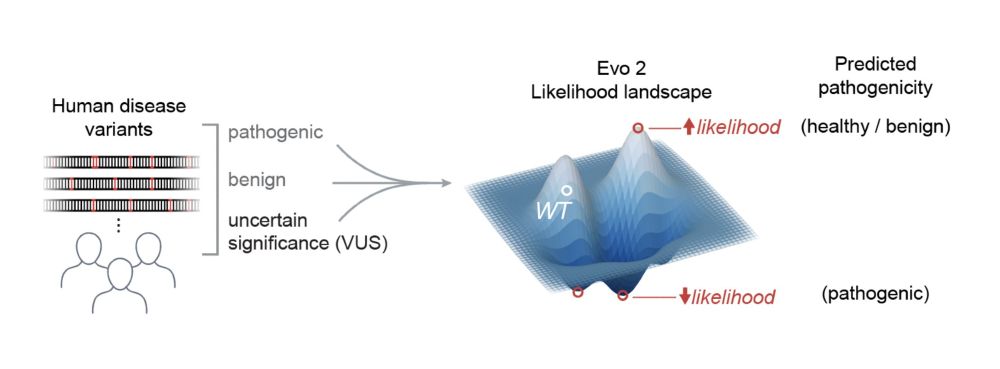
In other words, if you have a genetic mutation, Evo 2 has an opinion on whether or not it might cause disease


We release two models with 7B and 40B parameters along with weights, training and inference code, and pretraining data—making this one of the largest fully open AI models available
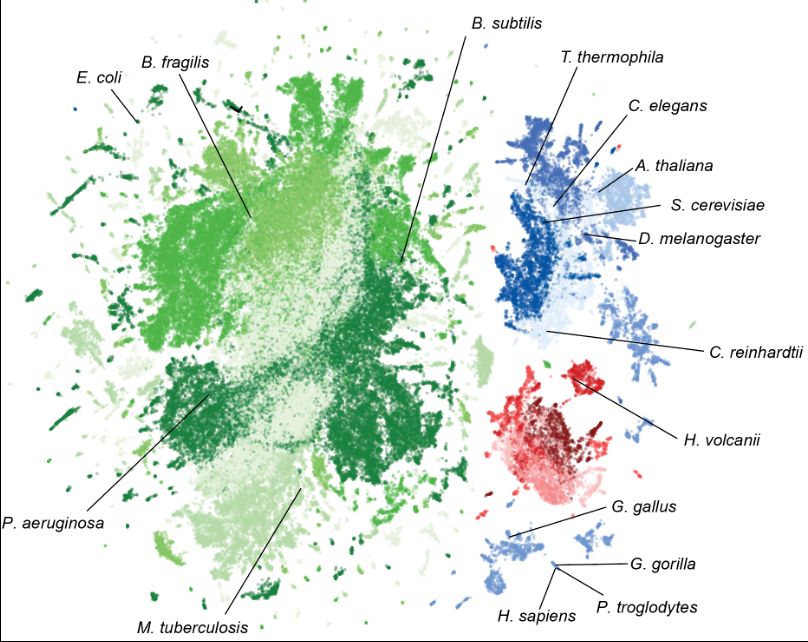
We release two models with 7B and 40B parameters along with weights, training and inference code, and pretraining data—making this one of the largest fully open AI models available
Today, @arcinstitute.org in collaboration with Nvidia releases Evo 2—a fully open source biological foundation model trained on genomes spanning the entire tree of life.
Today, @arcinstitute.org in collaboration with Nvidia releases Evo 2—a fully open source biological foundation model trained on genomes spanning the entire tree of life.

