If you want to develop new quantitative methods, collaborate across disciplines, and work on an exciting biological problem, we’d love to hear from you!
If you want to develop new quantitative methods, collaborate across disciplines, and work on an exciting biological problem, we’d love to hear from you!
The full project: centuri-livingsystems.org/wp-content/u...
The full project: centuri-livingsystems.org/wp-content/u...
academic.oup.com/nar/article/...

academic.oup.com/nar/article/...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
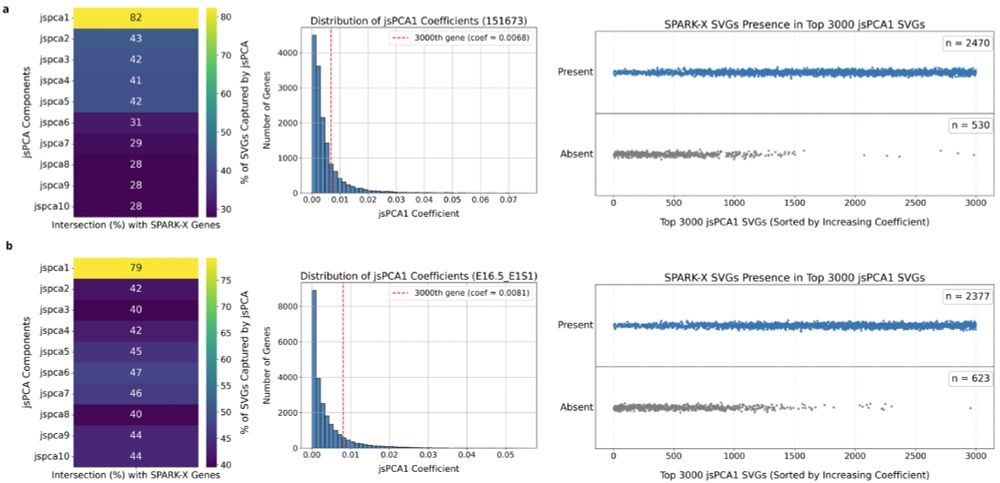
We have shown that it's fast, interpretable and highly adaptable to multiple settings and large datasets.
Congrats to Ines Assali who led the project!
We have shown that it's fast, interpretable and highly adaptable to multiple settings and large datasets.
Congrats to Ines Assali who led the project!
www.nature.com/articles/s41...
which was build on sPCA www.nature.com/articles/hdy....
To adapt it to spatial transcriptomics, we had to leverage sparsity and non convex optimization on manifold.

www.nature.com/articles/s41...
which was build on sPCA www.nature.com/articles/hdy....
To adapt it to spatial transcriptomics, we had to leverage sparsity and non convex optimization on manifold.