
Come do a PhD with us!
You will help us understand of how boundaries are established and maintained in early heart development.

Come do a PhD with us!
You will help us understand of how boundaries are established and maintained in early heart development.


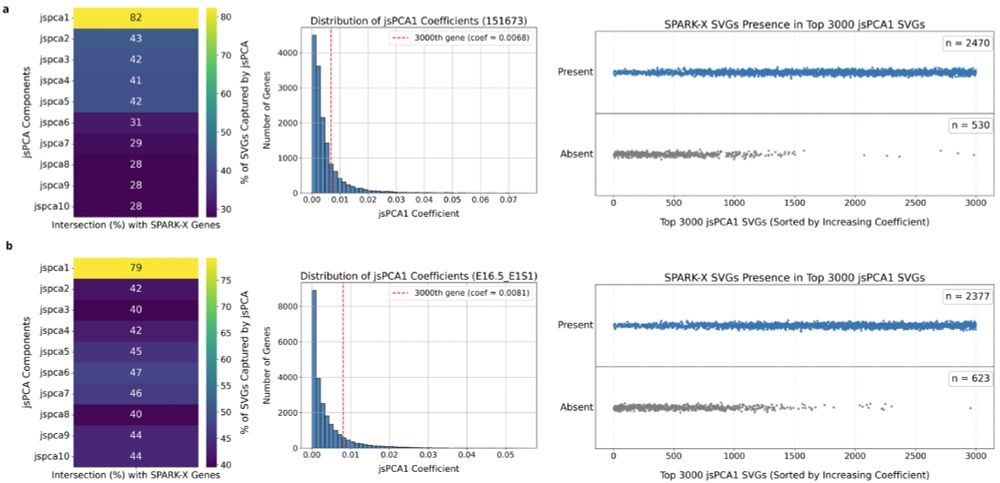











We solve a long-standing problem in the field: how to perform data integration for small unpaired datasets
Our solution: learning with class imbalance on both reference and target datasets!

We solve a long-standing problem in the field: how to perform data integration for small unpaired datasets
Our solution: learning with class imbalance on both reference and target datasets!
So many exciting mathematical approaches covering GRN inference, statistical testing and optimal transport.
Looking forward to the next edition!

So many exciting mathematical approaches covering GRN inference, statistical testing and optimal transport.
Looking forward to the next edition!
=> www.sciencedirect.com/science/arti...

=> www.sciencedirect.com/science/arti...
Preprint: arxiv.org/abs/2503.18856

Preprint: arxiv.org/abs/2503.18856



We proposed a new coupled variational auto-encoder architecture to solve the problem of data integration for small and unpaired datasets (diagonal integration)
arxiv.org/abs/2503.18856


We proposed a new coupled variational auto-encoder architecture to solve the problem of data integration for small and unpaired datasets (diagonal integration)
arxiv.org/abs/2503.18856

#SpatialOmics #SpatialBiology #Omics

#SpatialOmics #SpatialBiology #Omics






