
Paul Robustelli
@paulrobustelli.bsky.social
Assistant Professor at Dartmouth College
Computational Biophysics / Disordered Proteins / Molecular Recognition
Computational Biophysics / Disordered Proteins / Molecular Recognition
Pinned
Excited to share a new preprint:
"Monomer binding modes of small molecules that modulate the kinetics of hIAPP amyloid formation"
by graduate student Michelle Garcia together with post-doc Korey Reid.
Paper:
www.biorxiv.org/content/10.1...
Code + Ensembles: github.com/paulrobustel...
"Monomer binding modes of small molecules that modulate the kinetics of hIAPP amyloid formation"
by graduate student Michelle Garcia together with post-doc Korey Reid.
Paper:
www.biorxiv.org/content/10.1...
Code + Ensembles: github.com/paulrobustel...
Does anyone else think saying "this paper isn't a good fit for our journal" is quite a bit less silly than saying that a paper isn't a "striking advance" / "sufficiently timeless" / etc. ?.
Who is this goofy form letter language for exactly?
Who is this goofy form letter language for exactly?
November 1, 2025 at 3:24 PM
Does anyone else think saying "this paper isn't a good fit for our journal" is quite a bit less silly than saying that a paper isn't a "striking advance" / "sufficiently timeless" / etc. ?.
Who is this goofy form letter language for exactly?
Who is this goofy form letter language for exactly?
Happy to have contributed a few sections on NMR to this very practical and comprehensive community review on benchmarking force fields against experimental data.
In the latest @livecomsjournal.bsky.social
perpetual review, Cavender et al overview NMR and crystallographic experimental datasets that can be used to benchmark protein force fields, including best practices for setup and analysis of simulations!:
livecomsjournal.org/index.php/li...
#compchem
perpetual review, Cavender et al overview NMR and crystallographic experimental datasets that can be used to benchmark protein force fields, including best practices for setup and analysis of simulations!:
livecomsjournal.org/index.php/li...
#compchem

October 29, 2025 at 8:26 PM
Happy to have contributed a few sections on NMR to this very practical and comprehensive community review on benchmarking force fields against experimental data.
Today Writhe, our favorite knot theory descriptor for characterizing IDP dynamics, is the star of the lab pumpkin for the annual department carving contest.
If you can make sense of the design— I’ll buy you a coffee, bao bun, or wheatgrass shot at the next conference. No substitutions.
If you can make sense of the design— I’ll buy you a coffee, bao bun, or wheatgrass shot at the next conference. No substitutions.



October 23, 2025 at 1:05 AM
Today Writhe, our favorite knot theory descriptor for characterizing IDP dynamics, is the star of the lab pumpkin for the annual department carving contest.
If you can make sense of the design— I’ll buy you a coffee, bao bun, or wheatgrass shot at the next conference. No substitutions.
If you can make sense of the design— I’ll buy you a coffee, bao bun, or wheatgrass shot at the next conference. No substitutions.
Fun day at @dartmouthchem.bsky.social
serving buldak ramen and explaining the biophysics and structural biology of heat and capsaicin activation of the ion channels to celebrate @acs.org #NationalChemistryWeek.
serving buldak ramen and explaining the biophysics and structural biology of heat and capsaicin activation of the ion channels to celebrate @acs.org #NationalChemistryWeek.

October 20, 2025 at 6:52 PM
Fun day at @dartmouthchem.bsky.social
serving buldak ramen and explaining the biophysics and structural biology of heat and capsaicin activation of the ion channels to celebrate @acs.org #NationalChemistryWeek.
serving buldak ramen and explaining the biophysics and structural biology of heat and capsaicin activation of the ion channels to celebrate @acs.org #NationalChemistryWeek.
Our work developing a maximum entropy reweighting method to refine all-atom ensembles of IDPs with extensive NMR and SAXS datasets is now out in @natcomms.nature.com:
rdcu.be/eKlK7
Led by @dartmouthchem.bsky.social graduate student Kaushilk Borthakur in collaboration with @bonomimax.bsky.social
rdcu.be/eKlK7
Led by @dartmouthchem.bsky.social graduate student Kaushilk Borthakur in collaboration with @bonomimax.bsky.social

Determining accurate conformational ensembles of intrinsically disordered proteins at atomic resolution
Nature Communications - This study demonstrates how to combine molecular dynamics computer simulations with experimental biophysical data to determine accurate atomic-resolution ensembles of...
rdcu.be
October 10, 2025 at 3:33 PM
Our work developing a maximum entropy reweighting method to refine all-atom ensembles of IDPs with extensive NMR and SAXS datasets is now out in @natcomms.nature.com:
rdcu.be/eKlK7
Led by @dartmouthchem.bsky.social graduate student Kaushilk Borthakur in collaboration with @bonomimax.bsky.social
rdcu.be/eKlK7
Led by @dartmouthchem.bsky.social graduate student Kaushilk Borthakur in collaboration with @bonomimax.bsky.social
Excited to share a new preprint:
"Monomer binding modes of small molecules that modulate the kinetics of hIAPP amyloid formation"
by graduate student Michelle Garcia together with post-doc Korey Reid.
Paper:
www.biorxiv.org/content/10.1...
Code + Ensembles: github.com/paulrobustel...
"Monomer binding modes of small molecules that modulate the kinetics of hIAPP amyloid formation"
by graduate student Michelle Garcia together with post-doc Korey Reid.
Paper:
www.biorxiv.org/content/10.1...
Code + Ensembles: github.com/paulrobustel...
September 27, 2025 at 1:45 PM
Excited to share a new preprint:
"Monomer binding modes of small molecules that modulate the kinetics of hIAPP amyloid formation"
by graduate student Michelle Garcia together with post-doc Korey Reid.
Paper:
www.biorxiv.org/content/10.1...
Code + Ensembles: github.com/paulrobustel...
"Monomer binding modes of small molecules that modulate the kinetics of hIAPP amyloid formation"
by graduate student Michelle Garcia together with post-doc Korey Reid.
Paper:
www.biorxiv.org/content/10.1...
Code + Ensembles: github.com/paulrobustel...
Want to run and analyze MD simulations of a disordered protein?
Check out our step-by-step guide (+ tutorial with code) for preparing, running and analyzing replica exchange solute tempering (REST2) simulations of IDPs with @gromacs.bsky.social and @plumed.org on arxiv!
arxiv.org/abs/2505.01860
Check out our step-by-step guide (+ tutorial with code) for preparing, running and analyzing replica exchange solute tempering (REST2) simulations of IDPs with @gromacs.bsky.social and @plumed.org on arxiv!
arxiv.org/abs/2505.01860

Performing all-atom molecular dynamics simulations of intrinsically disordered proteins with replica exchange solute tempering
All-atom molecular dynamics (MD) computer simulations are a valuable tool for characterizing the conformational ensembles of intrinsically disordered proteins (IDPs). IDP conformational ensembles are ...
arxiv.org
May 6, 2025 at 2:19 AM
Want to run and analyze MD simulations of a disordered protein?
Check out our step-by-step guide (+ tutorial with code) for preparing, running and analyzing replica exchange solute tempering (REST2) simulations of IDPs with @gromacs.bsky.social and @plumed.org on arxiv!
arxiv.org/abs/2505.01860
Check out our step-by-step guide (+ tutorial with code) for preparing, running and analyzing replica exchange solute tempering (REST2) simulations of IDPs with @gromacs.bsky.social and @plumed.org on arxiv!
arxiv.org/abs/2505.01860
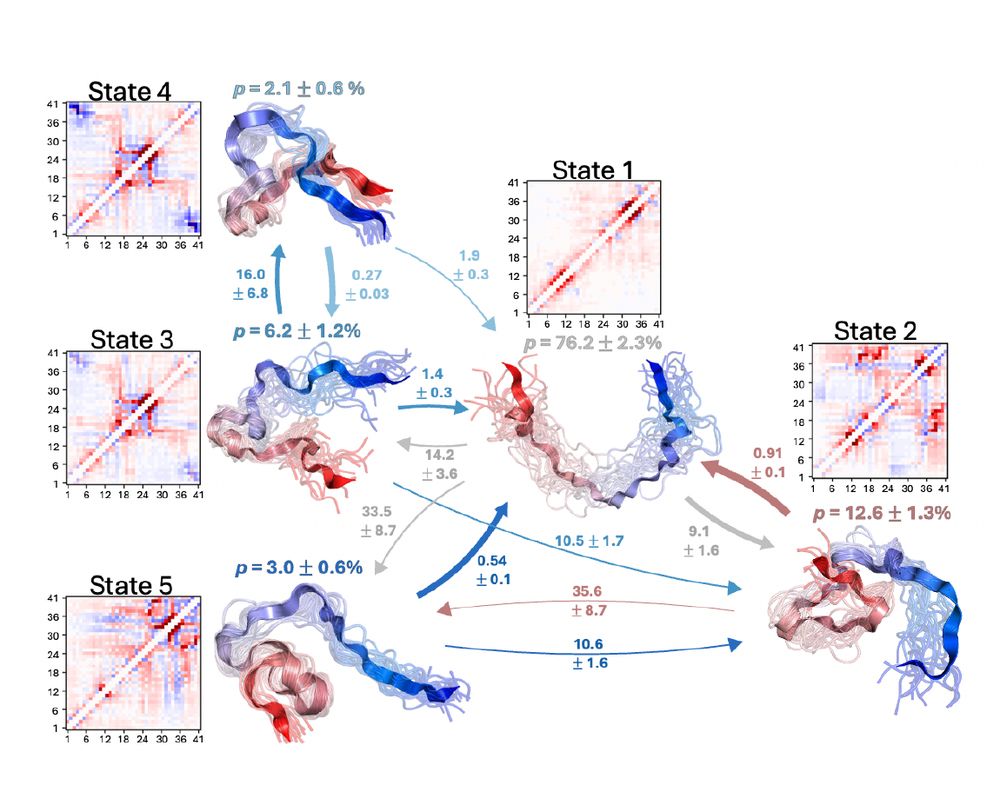
Presenting one of my favorite manuscripts I've ever worked on:
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
April 30, 2025 at 5:45 PM
Presenting one of my favorite manuscripts I've ever worked on:
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
Thanks to my hosts @bonomimax.bsky.social, @xsalvatella1.bsky.social, @fabferrage.bsky.social, @rcrehuet.bsky.social & Gabi Heller for a wonderful sabbatical science tour of Barcelona, Paris and London! Until next time!




February 27, 2025 at 3:51 PM
Thanks to my hosts @bonomimax.bsky.social, @xsalvatella1.bsky.social, @fabferrage.bsky.social, @rcrehuet.bsky.social & Gabi Heller for a wonderful sabbatical science tour of Barcelona, Paris and London! Until next time!
As an American who’s immigrant grandfather grew up in a New Jersey home with a dirt floor, I can’t quite explain the feeling I have when I walk past the pantheon to spend the afternoon discussing magnetic resonance with Parisian intellectuals.
Thanks @fabferrage.bsky.social for a great visit!
Thanks @fabferrage.bsky.social for a great visit!
We had a great visit of @paulrobustelli.bsky.social today at @chimieens.bsky.social. Inspiring new ideas in a beautiful seminar and lively discussions. The whole day went by too fast!

February 26, 2025 at 8:46 AM
As an American who’s immigrant grandfather grew up in a New Jersey home with a dirt floor, I can’t quite explain the feeling I have when I walk past the pantheon to spend the afternoon discussing magnetic resonance with Parisian intellectuals.
Thanks @fabferrage.bsky.social for a great visit!
Thanks @fabferrage.bsky.social for a great visit!
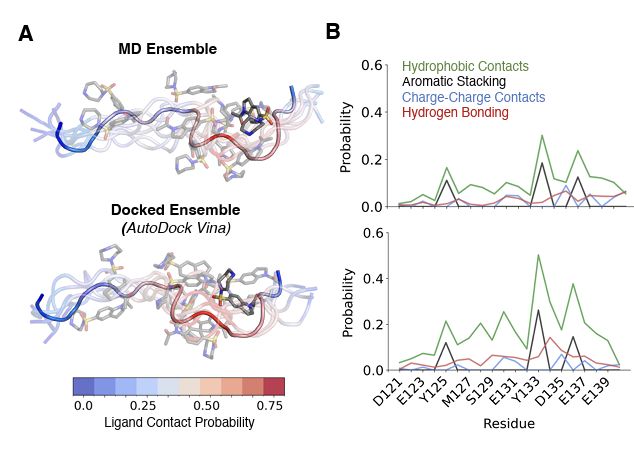
Proud to share:
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
January 31, 2025 at 7:59 PM
Proud to share:
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
"Ensemble docking for intrinsically disordered proteins"
from Dartmouth undergrad Anjali Dhar 24' and grad student Tommy Sisk. We present ensemble docking strategies for IDPs that, remarkably, seem to work!
www.biorxiv.org/content/10.1...
Code: github.com/paulrobustel...
One note about the DiffDock *preprint* fracas: it seems like two of the main issues at hand
i) experienced researchers insisting on addition controls to justify claims
ii) objections to overly combative language and potentially misleading comparisons
are things that peer reviewed journals...
i) experienced researchers insisting on addition controls to justify claims
ii) objections to overly combative language and potentially misleading comparisons
are things that peer reviewed journals...
December 10, 2024 at 5:49 PM
One note about the DiffDock *preprint* fracas: it seems like two of the main issues at hand
i) experienced researchers insisting on addition controls to justify claims
ii) objections to overly combative language and potentially misleading comparisons
are things that peer reviewed journals...
i) experienced researchers insisting on addition controls to justify claims
ii) objections to overly combative language and potentially misleading comparisons
are things that peer reviewed journals...
I’m looking for examples of the largest conformational changes that have been captured in ensemble docking or flexible docking approaches.
Can anyone point me to some good examples?
Can anyone point me to some good examples?
December 6, 2024 at 4:09 PM
I’m looking for examples of the largest conformational changes that have been captured in ensemble docking or flexible docking approaches.
Can anyone point me to some good examples?
Can anyone point me to some good examples?
I'm excited to share a new manuscript with Jiaqi Zhu characterizing the conformational ensembles of covalent adducts formed by a disordered protein and small molecule drugs:
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...
November 24, 2024 at 5:27 PM
I'm excited to share a new manuscript with Jiaqi Zhu characterizing the conformational ensembles of covalent adducts formed by a disordered protein and small molecule drugs:
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...



