Our new paper is just out in PNAS:
www.pnas.org/doi/10.1073/...
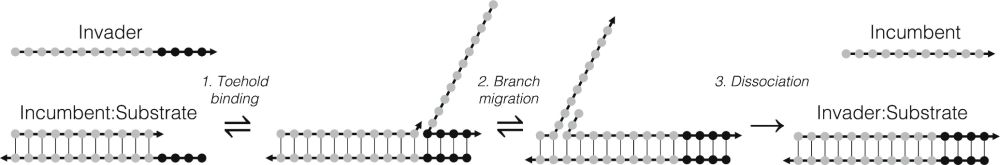
It is about nucleic acid strand displacement, which is a key reaction in molecular computation, and part of the success of the oxDNA model comes from the fact it can efficiently simulate this process.
1/4🧵
Our new paper is just out in PNAS:
www.pnas.org/doi/10.1073/...
It is about nucleic acid strand displacement, which is a key reaction in molecular computation, and part of the success of the oxDNA model comes from the fact it can efficiently simulate this process.
1/4🧵
biorxiv.org/content/10.1...
Excellent work by Rebecca, "Metrology4life" 2nd PhD student and Great collaboration with @petrsulc.bsky.social

onlinelibrary.wiley.com/doi/10.1002/...
onlinelibrary.wiley.com/doi/10.1002/...
doi.org/10.1145/3703...

doi.org/10.1145/3703...

