
Interested in computational chemistry in @orca-qc-official.bsky.social
Linux fan
orcaforum.kofo.mpg.de/app.php/dlex...
This is strictly a bugfix release that takes care of the problems that have been reported since the release of ORCA 6.1. The manual has also been updated.
orcaforum.kofo.mpg.de/app.php/dlex...
This is strictly a bugfix release that takes care of the problems that have been reported since the release of ORCA 6.1. The manual has also been updated.
doi.org/10.1002/cptc...
CASSCF in ORCA: www.faccts.de/docs/orca/6....
#ORCAqc #CompChem #QuantumChem

doi.org/10.1002/cptc...
CASSCF in ORCA: www.faccts.de/docs/orca/6....
#ORCAqc #CompChem #QuantumChem
Find the best time for your group chat to hang out, choose a vacation destination, or ask if you should give yourself a haircut at 1 AM. Now in the latest version of Signal for iOS, Android, and Desktop.
signal.org/blog/polls/

Find the best time for your group chat to hang out, choose a vacation destination, or ask if you should give yourself a haircut at 1 AM. Now in the latest version of Signal for iOS, Android, and Desktop.
signal.org/blog/polls/
#ORCAqc #ORCA6 #massspectrometry #CID #CompChem #ChemSky #CompChemSky
Just published in #JASMS : doi.org/10.1021/jasms.5c00234
Grateful to my coauthors Stefan Grimme @grimmelab.bsky.social & Marianne Engeser @unibonn.bsky.social - this is the last project of my PhD and completes my work on QCxMS2!
#MassSpec #compchem
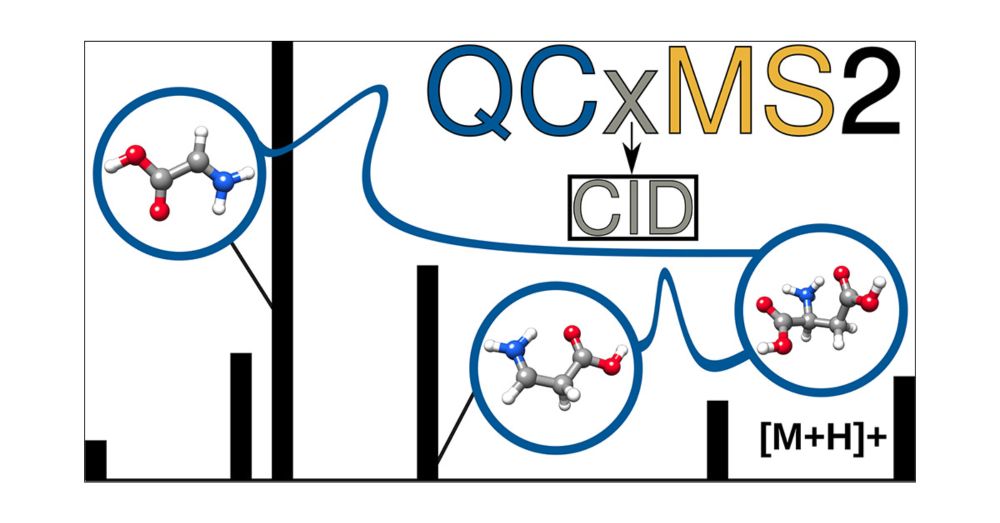
#ORCAqc #ORCA6 #massspectrometry #CID #CompChem #ChemSky #CompChemSky
doi.org/10.1021/acs....
#ORCA #ORCAqc #CompChem #QuantumChem #TheoChem #CASSCF

doi.org/10.1021/acs....
#ORCA #ORCAqc #CompChem #QuantumChem #TheoChem #CASSCF

