(he/him)
Just published in #JASMS : doi.org/10.1021/jasms.5c00234
Grateful to my coauthors Stefan Grimme @grimmelab.bsky.social & Marianne Engeser @unibonn.bsky.social - this is the last project of my PhD and completes my work on QCxMS2!
#MassSpec #compchem
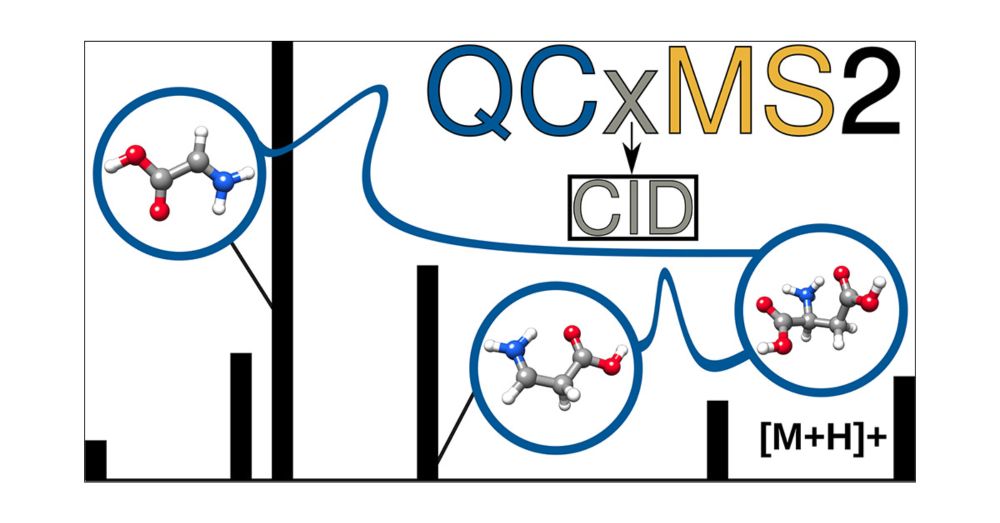
Just published in #JASMS : doi.org/10.1021/jasms.5c00234
Grateful to my coauthors Stefan Grimme @grimmelab.bsky.social & Marianne Engeser @unibonn.bsky.social - this is the last project of my PhD and completes my work on QCxMS2!
#MassSpec #compchem
Catch the details at #WATOC: my talk (Thu Session B1) and Stefan’s talk (Thu Session A2).
#compchem
doi.org/10.26434/che...

Catch the details at #WATOC: my talk (Thu Session B1) and Stefan’s talk (Thu Session A2).
#compchem
doi.org/10.26434/che...
#CompChemSky #MassSpecSky
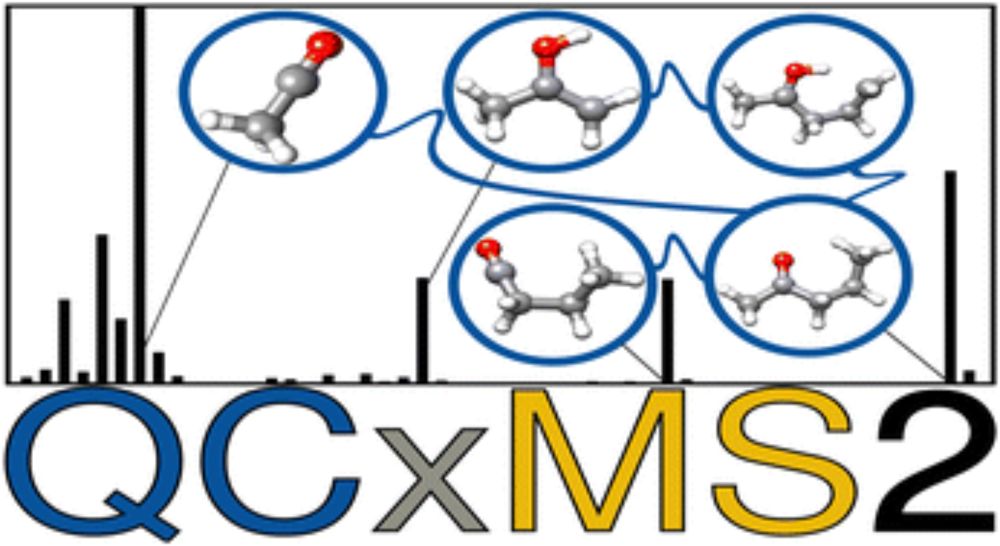
#CompChemSky #MassSpecSky
Check out our next-generation mass spectra calculation program for EI-MS, based on automated reaction network discovery @grimmelab.bsky.social
-preprint @chemrxiv.bsky.social doi.org/10.26434/che...
-software available at github.com/grimme-lab/Q...
#Massspec #Compchem

Check out our next-generation mass spectra calculation program for EI-MS, based on automated reaction network discovery @grimmelab.bsky.social
-preprint @chemrxiv.bsky.social doi.org/10.26434/che...
-software available at github.com/grimme-lab/Q...
#Massspec #Compchem
Learn more at https://buff.ly/3BXDVhO.
#ChemistryAward #ChemistryEurope

Learn more at https://buff.ly/3BXDVhO.
#ChemistryAward #ChemistryEurope
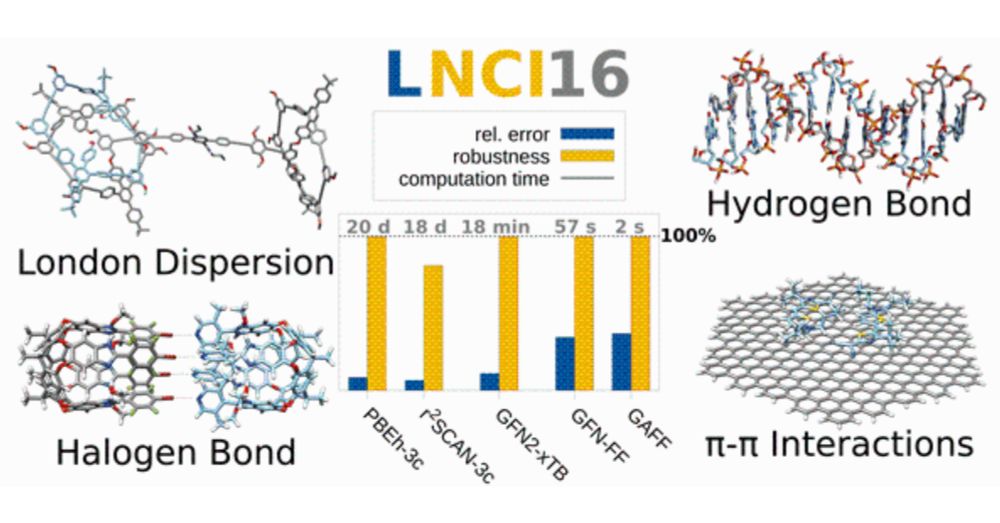
and see how well CREST and CENSO work for this challenging task: doi.org/10.1039/D2CP... @grimmelab.bsky.social
#compchem
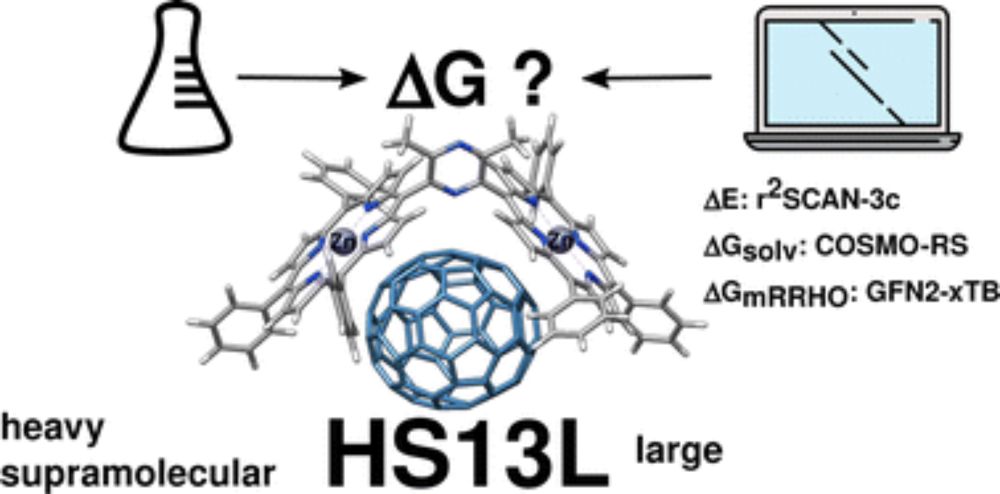
and see how well CREST and CENSO work for this challenging task: doi.org/10.1039/D2CP... @grimmelab.bsky.social
#compchem
#compchem #research

#compchem #research

