
Neel Shah
@nshahlab.bsky.social
Assistant Professor at Columbia Chemistry. PI of a chemical biology lab full of awesome people. Internal conflicts: Chemist or biologist? Kinase or phosphatase?
Lab website: https://shahlab.wixsite.com/home
ORCiD: 0000-0002-1186-0626
Lab website: https://shahlab.wixsite.com/home
ORCiD: 0000-0002-1186-0626
Had a great lab retreat today in Central Park, with team-building games, great conversations, and lots of laughs, followed by delicious food/drinks. Gearing up for my State of the Lab address tmrw, where I’ll recap our past year and discuss what’s on the horizon! So lucky to have this awesome group!

August 1, 2025 at 12:41 AM
Had a great lab retreat today in Central Park, with team-building games, great conversations, and lots of laughs, followed by delicious food/drinks. Gearing up for my State of the Lab address tmrw, where I’ll recap our past year and discuss what’s on the horizon! So lucky to have this awesome group!
Congrats to PhD #5 from our lab, Dr. Anne van Vlimmeren, who successfully defended her dissertation yesterday!!! Check out some of Anne's thesis work on the diverse structural/functional consequences of SHP2 mutations: PMIDs 39012820, 40661614, 40631144, 40631229.
July 22, 2025 at 11:28 PM
Congrats to PhD #5 from our lab, Dr. Anne van Vlimmeren, who successfully defended her dissertation yesterday!!! Check out some of Anne's thesis work on the diverse structural/functional consequences of SHP2 mutations: PMIDs 39012820, 40661614, 40631144, 40631229.
Congratulations to Dr. Ziyuan (Jason) Jiang, the 4th PhD from our lab! He gave a stellar thesis defense presentation on SHP2 deep mutational scanning, structure, and dynamics. We even managed to squeeze in a new lab photo in the midst of all of the excitement and celebrations!




July 9, 2025 at 1:47 AM
Congratulations to Dr. Ziyuan (Jason) Jiang, the 4th PhD from our lab! He gave a stellar thesis defense presentation on SHP2 deep mutational scanning, structure, and dynamics. We even managed to squeeze in a new lab photo in the midst of all of the excitement and celebrations!
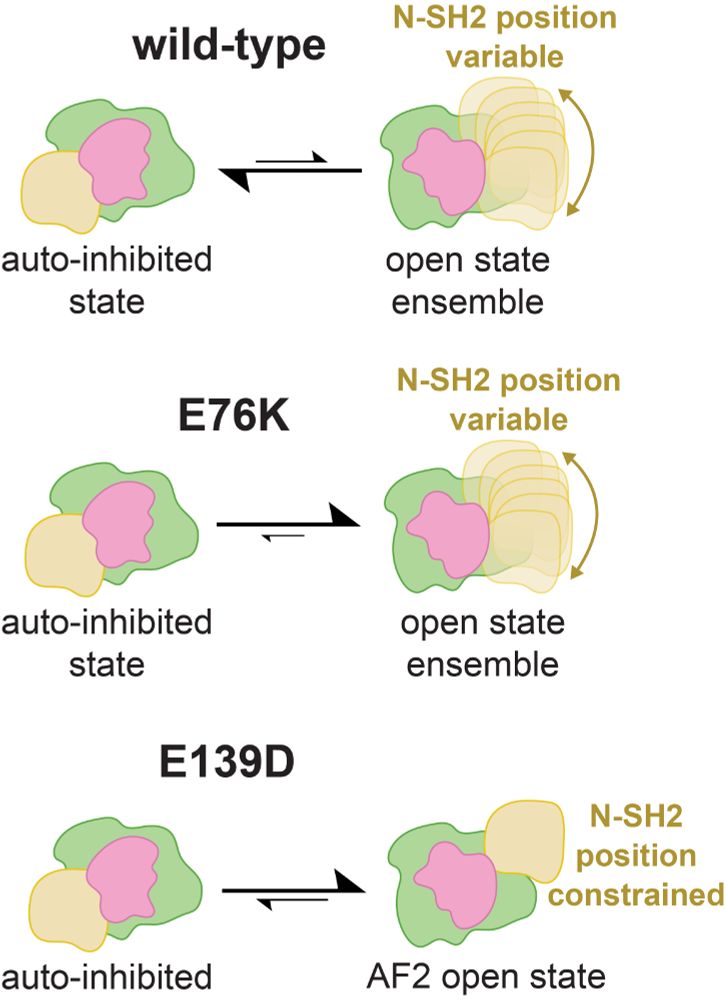
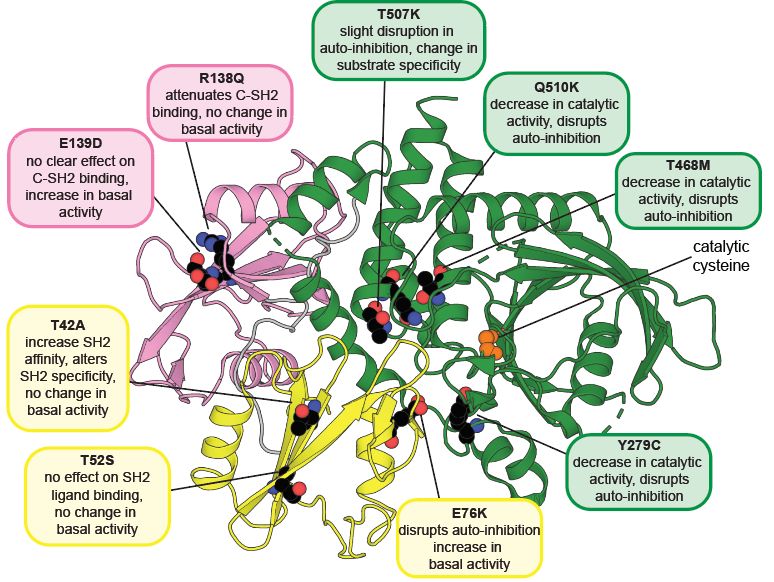
A neat upshot of this mechanism is that, unlike other activating mutations, which destabilize auto-inhibited SHP2 and yield an ensemble of active states, the active state of E139D is more conformationally constrained. Ultimately, this impacts the phosphoprotein binding specificity of this mutant.
July 5, 2025 at 4:58 PM
A neat upshot of this mechanism is that, unlike other activating mutations, which destabilize auto-inhibited SHP2 and yield an ensemble of active states, the active state of E139D is more conformationally constrained. Ultimately, this impacts the phosphoprotein binding specificity of this mutant.
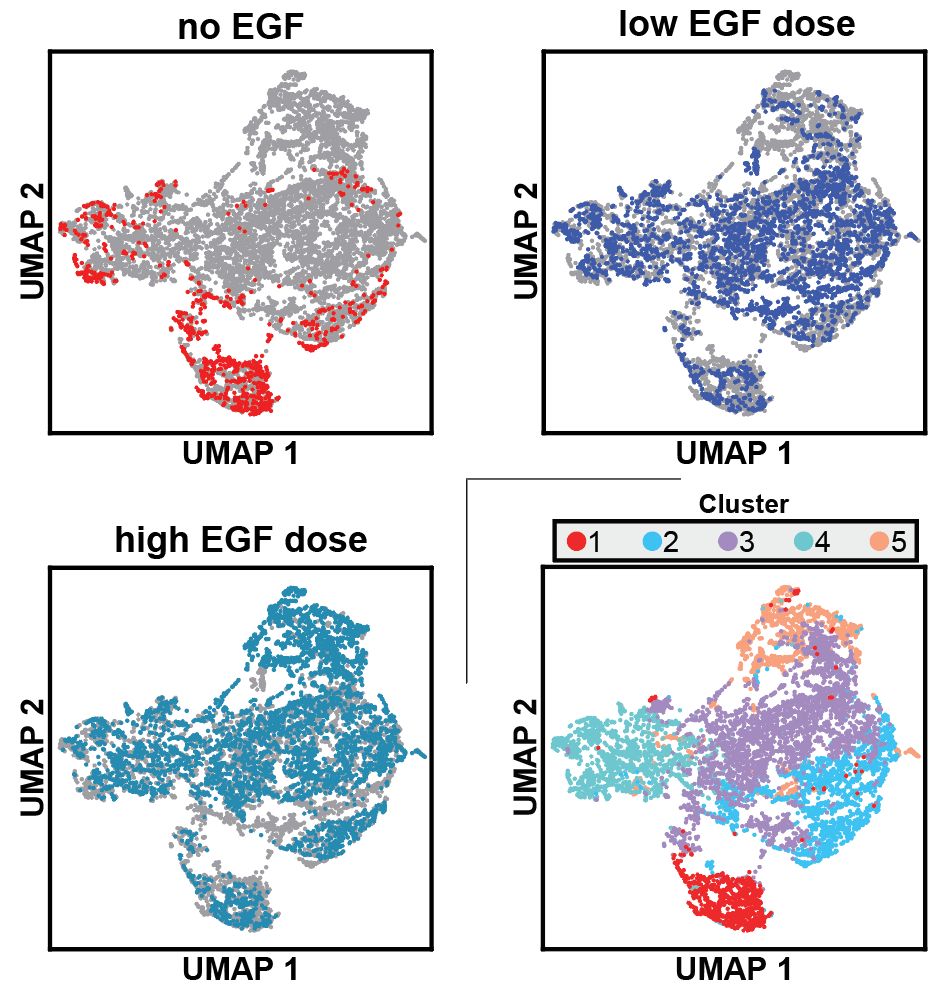
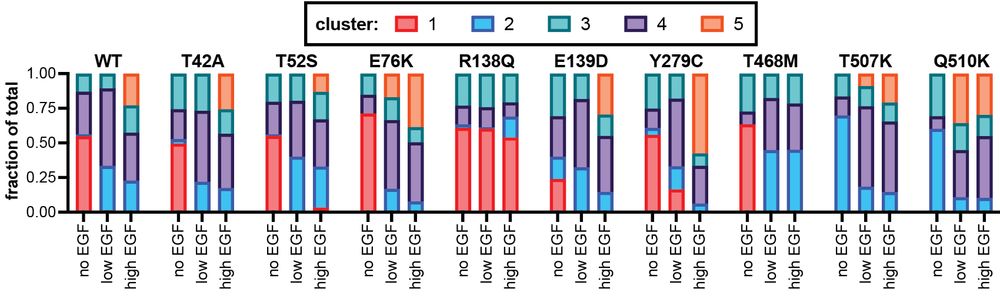
Through some data analysis (read the paper!) we identified 5 distinct clusters of cell states and found that different mutants occupy different cell states as a function of EGF stimulation
July 3, 2025 at 5:31 PM
Through some data analysis (read the paper!) we identified 5 distinct clusters of cell states and found that different mutants occupy different cell states as a function of EGF stimulation
Through this experiment, we first identified a core SHP2-dependent transcriptome and identified key EGF-response programs that are SHP2 dependent.
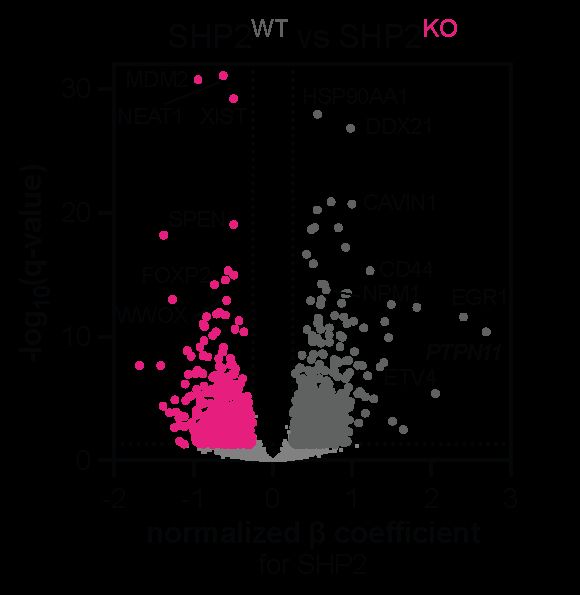
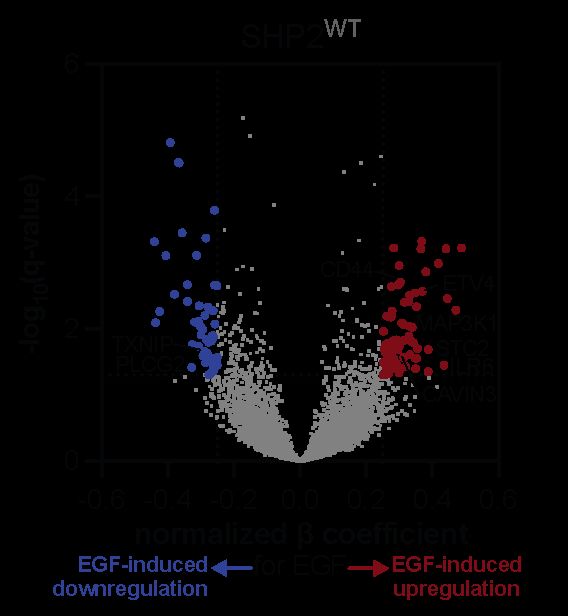
July 3, 2025 at 5:31 PM
Through this experiment, we first identified a core SHP2-dependent transcriptome and identified key EGF-response programs that are SHP2 dependent.
To get at this question, Anne and Ross designed as simple but elegant experiment, where they reconstituted SHP2 knock-out HEK293 cells with different SHP2 variants, stimulated with EGF for different doses/timepoints, and then did multiplexed single cell transcriptomics.
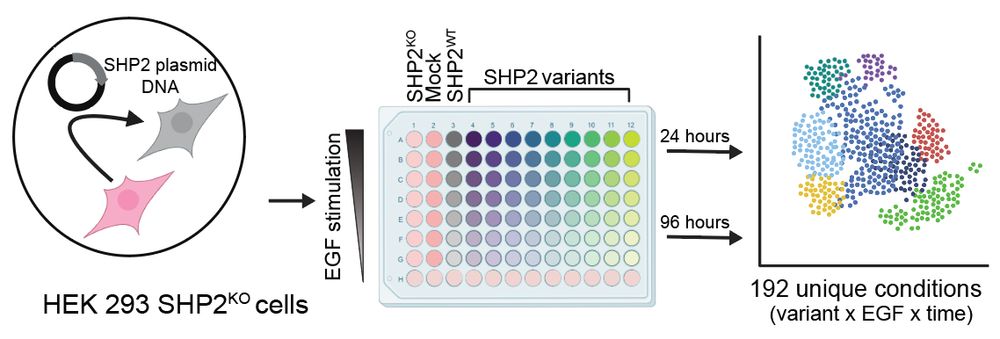
July 3, 2025 at 5:31 PM
To get at this question, Anne and Ross designed as simple but elegant experiment, where they reconstituted SHP2 knock-out HEK293 cells with different SHP2 variants, stimulated with EGF for different doses/timepoints, and then did multiplexed single cell transcriptomics.
SHP2 is mutated in developmental disorders and cancers. We are (compulsively) focused on understanding the diverse ways that mutations dysregulate this enzyme: changes in catalytic activity, dynamics, stability, interactions, etc. But do these different mechanism yield different downstream outcomes?
July 3, 2025 at 5:31 PM
SHP2 is mutated in developmental disorders and cancers. We are (compulsively) focused on understanding the diverse ways that mutations dysregulate this enzyme: changes in catalytic activity, dynamics, stability, interactions, etc. But do these different mechanism yield different downstream outcomes?
Focusing on happy news… I’m so proud of our undergrad researcher Zijing for her graduation from Barnard this week. Zijing did superb work on kinase and SH2 sequence recognition while in the lab (papers coming in the next year). I’m also happy to announce the awesome award I just received (from her).


May 23, 2025 at 6:35 PM
Focusing on happy news… I’m so proud of our undergrad researcher Zijing for her graduation from Barnard this week. Zijing did superb work on kinase and SH2 sequence recognition while in the lab (papers coming in the next year). I’m also happy to announce the awesome award I just received (from her).
Something positive: Nice weather and overdue celebrations with the lab for a paper accepted, another preprinted, two qualifying exams finished, a recent grant (?!), and general awesomeness, of course!

May 2, 2025 at 11:31 PM
Something positive: Nice weather and overdue celebrations with the lab for a paper accepted, another preprinted, two qualifying exams finished, a recent grant (?!), and general awesomeness, of course!
I love science, I love being a scientist, and I love supporting and training scientists who will go on to make awesome discoveries and push the frontiers of knowledge to benefit society. I will always Stand Up for Science, and so should you!


March 7, 2025 at 8:16 PM
I love science, I love being a scientist, and I love supporting and training scientists who will go on to make awesome discoveries and push the frontiers of knowledge to benefit society. I will always Stand Up for Science, and so should you!
Finally, we examined proximity labeling in the presence of the clinically-relevant inhibitor TNO155. This inhibitor stabilizes the autoinhibited state of SHP2, which should prevent some protein-protein interactions. Indeed we see reduced proximity labeling when the inhibitor is present.

March 3, 2025 at 5:16 PM
Finally, we examined proximity labeling in the presence of the clinically-relevant inhibitor TNO155. This inhibitor stabilizes the autoinhibited state of SHP2, which should prevent some protein-protein interactions. Indeed we see reduced proximity labeling when the inhibitor is present.
We spent some time exploring how SHP2 mutants might get in to the mitochondria more than the wild-type enzyme. Too much to unpack in a Bluetorial, but we think it has to do with protein stability, interaction with mitochondrial import machinery, and mitochondrial chaperones.



March 3, 2025 at 5:16 PM
We spent some time exploring how SHP2 mutants might get in to the mitochondria more than the wild-type enzyme. Too much to unpack in a Bluetorial, but we think it has to do with protein stability, interaction with mitochondrial import machinery, and mitochondrial chaperones.
We were also able to use our SH2 predictions to identify potential SHP2 binding sites on mitochondrial proteins. We validated one of these predicted binders: ATPAF1, which is an assembly factor for the F1 component of the mitochondrial ATP synthase.


March 3, 2025 at 5:16 PM
We were also able to use our SH2 predictions to identify potential SHP2 binding sites on mitochondrial proteins. We validated one of these predicted binders: ATPAF1, which is an assembly factor for the F1 component of the mitochondrial ATP synthase.
Perhaps one of the coolest things we found is that select SHP2 mutants have enhanced mitochondrial signal in the TurboID experiments (over TurboID-only negative controls and over wild-type SHP2), which we validated via mito isolations. In particular, we see enhanced association with ETC proteins.


March 3, 2025 at 5:16 PM
Perhaps one of the coolest things we found is that select SHP2 mutants have enhanced mitochondrial signal in the TurboID experiments (over TurboID-only negative controls and over wild-type SHP2), which we validated via mito isolations. In particular, we see enhanced association with ETC proteins.
Things got very interesting when we dug deeper into the data for WT and mutants and found lots of mitochondrial proteins. There are scattered reports of SHP2 in the mitochondria, in some cases impacting respiration. And some mutant-SHP2-driven congenital disorders resemble mitochondrial disorders.

March 3, 2025 at 5:16 PM
Things got very interesting when we dug deeper into the data for WT and mutants and found lots of mitochondrial proteins. There are scattered reports of SHP2 in the mitochondria, in some cases impacting respiration. And some mutant-SHP2-driven congenital disorders resemble mitochondrial disorders.
For example, for T42A, a mutant that we previously showed enhances SH2 binding affinity and alters sequence specificity, we saw increased TurboID signal, and proteins with enhanced signal also had phosphosites with enhanced affinity for the mutant in our peptide display screens.

March 3, 2025 at 5:16 PM
For example, for T42A, a mutant that we previously showed enhances SH2 binding affinity and alters sequence specificity, we saw increased TurboID signal, and proteins with enhanced signal also had phosphosites with enhanced affinity for the mutant in our peptide display screens.
One thing we did throughout the paper was juxtapose our proximity-labeling data with our SHP2 SH2 domain specificity profiling via peptide display, some old data (PMIDs 36927728 and 39012820) and some new data. Our specificity profiling method is pretty good at predicting known SHP2 SH2 interactors.

March 3, 2025 at 5:16 PM
One thing we did throughout the paper was juxtapose our proximity-labeling data with our SHP2 SH2 domain specificity profiling via peptide display, some old data (PMIDs 36927728 and 39012820) and some new data. Our specificity profiling method is pretty good at predicting known SHP2 SH2 interactors.
We did a lot of quality-control experiments to be sure we were detecting real signal on functional SHP2 molecules (see SI). Our WT datasets were comforting in these regards, highlighting plenty of known interactors:

March 3, 2025 at 5:16 PM
We did a lot of quality-control experiments to be sure we were detecting real signal on functional SHP2 molecules (see SI). Our WT datasets were comforting in these regards, highlighting plenty of known interactors:
Most tyrosine phosphatase protein-protein interactions are transient, and not surprisingly, we failed to pick up much in affinity purification-MS experiments. So we turned to TurboID proximity labeling on wild-type SHP2, 10 disease mutants, and an inhibitor-bound state.

March 3, 2025 at 5:16 PM
Most tyrosine phosphatase protein-protein interactions are transient, and not surprisingly, we failed to pick up much in affinity purification-MS experiments. So we turned to TurboID proximity labeling on wild-type SHP2, 10 disease mutants, and an inhibitor-bound state.
SHP2 mutations often induce large conformational changes that expose binding interfaces, or mutations are at key interfaces and remodel their affinity/specificity. As such, we hypothesized that mutations are likely to have diverse effects on protein-protein interactions.

March 3, 2025 at 5:16 PM
SHP2 mutations often induce large conformational changes that expose binding interfaces, or mutations are at key interfaces and remodel their affinity/specificity. As such, we hypothesized that mutations are likely to have diverse effects on protein-protein interactions.
We have been intrigued for a while by the plethora of disease-associated mutations in SHP2, and their distinct biochemical and structural properties. Much of the cell signaling that underlies differences in the pathogenicity of these mutants remains murky.

March 3, 2025 at 5:16 PM
We have been intrigued for a while by the plethora of disease-associated mutations in SHP2, and their distinct biochemical and structural properties. Much of the cell signaling that underlies differences in the pathogenicity of these mutants remains murky.
This job has so many ups and downs, but one constant that I’m always thankful for is this creative, hard-working, supportive group (they support me, too!!). I’m also thankful for all the awesome ideas and cool data they generate, which keep us all motivated and stimulated and make this so much fun!

November 28, 2024 at 1:41 PM
This job has so many ups and downs, but one constant that I’m always thankful for is this creative, hard-working, supportive group (they support me, too!!). I’m also thankful for all the awesome ideas and cool data they generate, which keep us all motivated and stimulated and make this so much fun!
Overall, we find about a half dozen different ways that clinical mutations can dysregulate SHP2 (both gain and loss of function) and take a deep dive into the structural and functional consequences of each one. We further explore the mechanisms using MD, biochem, biophysical, and cellular assays.

November 20, 2024 at 12:50 PM
Overall, we find about a half dozen different ways that clinical mutations can dysregulate SHP2 (both gain and loss of function) and take a deep dive into the structural and functional consequences of each one. We further explore the mechanisms using MD, biochem, biophysical, and cellular assays.
Juxtaposition of the full length and isolated phosphatase domain datasets, along with clinical variant databases, reveals a broad range of mutational effects that dysregulate SHP2 activity in different ways in different human diseases.

November 20, 2024 at 12:50 PM
Juxtaposition of the full length and isolated phosphatase domain datasets, along with clinical variant databases, reveals a broad range of mutational effects that dysregulate SHP2 activity in different ways in different human diseases.

