https://nsapoval.github.io/
#singlecell #GRN #LLM

#singlecell #GRN #LLM

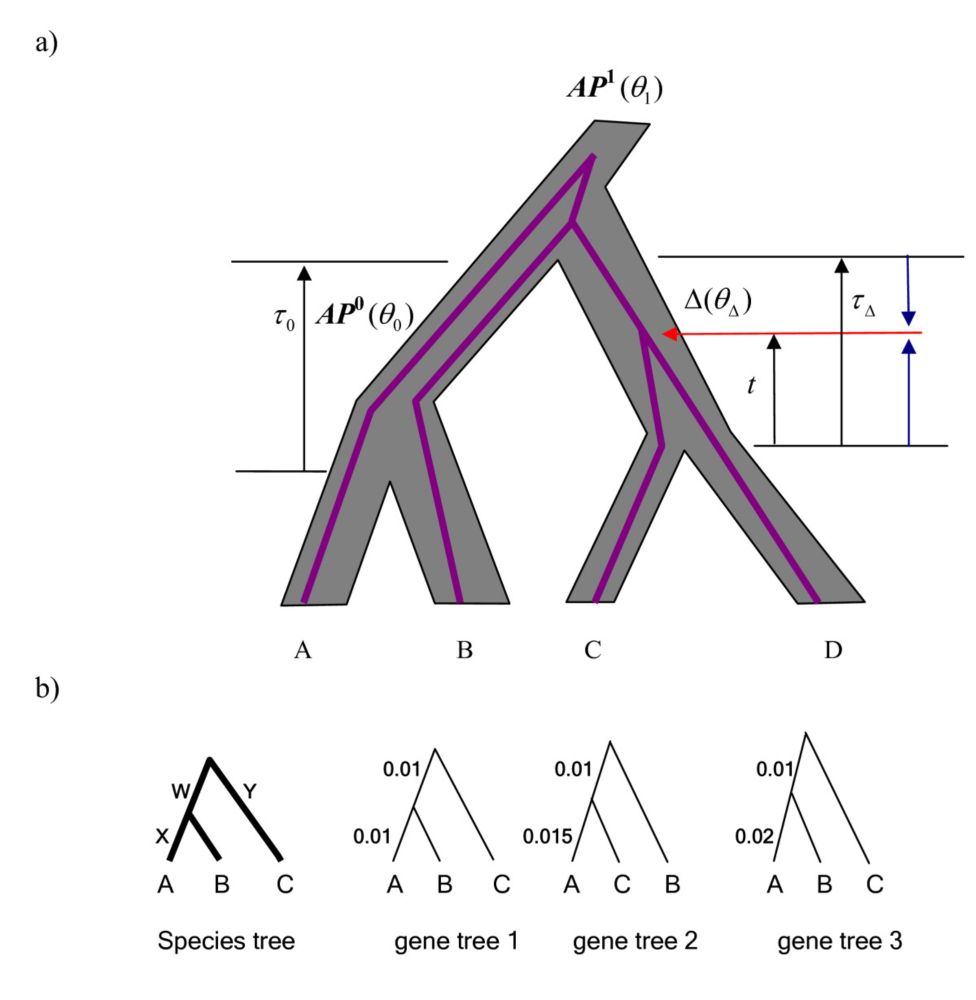
A fantastic theory talk, offering intuitive insights!
Paper: doi.org/10.1101/2025.01.28.635282



A fantastic theory talk, offering intuitive insights!
Paper: doi.org/10.1101/2025.01.28.635282