
Working at the intersection of functional genomics, systems biology, and machine learning. I also build rusty bioinformatics tools
https://github.com/noamteyssier


This lets you skip an intermediary write step and go straight from SRA to downstream tools.
It works with accessions that are both on- or off-disk.

This lets you skip an intermediary write step and go straight from SRA to downstream tools.
It works with accessions that are both on- or off-disk.
But you're right I didn't formally test it. Here's a simple bench with Kent's utils (1-core bqtools to be fair)


But you're right I didn't formally test it. Here's a simple bench with Kent's utils (1-core bqtools to be fair)
I forked sassy for a quick test and found 25x throughput over gzip and a 16x over raw.
That includes sharing SIMD with search during 2bit decode also.

I forked sassy for a quick test and found 25x throughput over gzip and a 16x over raw.
That includes sharing SIMD with search during 2bit decode also.
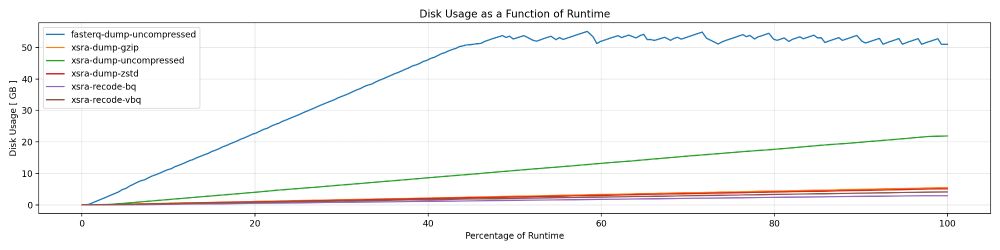
It can achieve almost 10x faster throughput outputting BINSEQ files when skipping FASTQ altogether

It can achieve almost 10x faster throughput outputting BINSEQ files when skipping FASTQ altogether


We provide rust libraries for IO, C and C++ bindings to BINSEQ, and a CLI tool to easily manipulate them.

We provide rust libraries for IO, C and C++ bindings to BINSEQ, and a CLI tool to easily manipulate them.

