Currently: Stats PhD @Harvard
Previously: CS/Soc @Stanford, Stat/ML @Oxford
https://njw.fish
GDEs offer a general framework for:
1. Modeling hierarchical data
2. Embedding and generating distributions
3. Linking modern generative models with classical statistical ideas
📄 arxiv.org/abs/2505.18150
💻 github.com/njwfish/Dist...

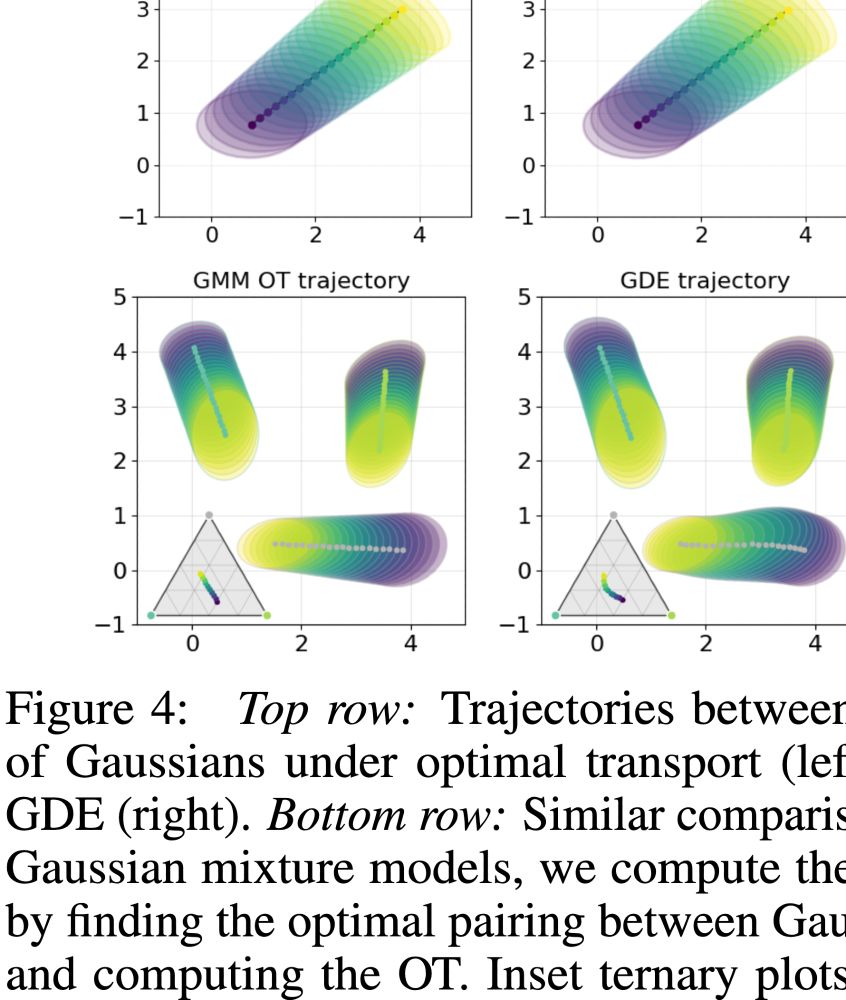
GDE latent distances track Wasserstein-2 (W₂) distances across modalities (shown for multinomial distributions)
Latent interpolations recover optimal transport paths (shown for Gaussians and Gaussian mixtures)

GDEs offer a general framework for:
1. Modeling hierarchical data
2. Embedding and generating distributions
3. Linking modern generative models with classical statistical ideas
📄 arxiv.org/abs/2505.18150
💻 github.com/njwfish/Dist...

We’re excited about GDEs as a tool for multiscale modeling — and as a bridge between empirical process theory, generative modeling, and geometry.
Looking forward to your thoughts!
We’re excited about GDEs as a tool for multiscale modeling — and as a bridge between empirical process theory, generative modeling, and geometry.
Looking forward to your thoughts!
GDEs offer a general framework for:
1. Modeling hierarchical data
2. Embedding and generating distributions
3. Linking modern generative models with classical statistical ideas
📄 arxiv.org/abs/2505.18150
💻 github.com/njwfish/Dist...

GDEs offer a general framework for:
1. Modeling hierarchical data
2. Embedding and generating distributions
3. Linking modern generative models with classical statistical ideas
📄 arxiv.org/abs/2505.18150
💻 github.com/njwfish/Dist...
Sequences binned by expression → GDEs provide a powerful embedding space for regulatory sequence design.
🦠 SARS-CoV-2 spike protein dynamics
GDEs embed monthly lineage distributions and recover smooth latent chronologies.


Sequences binned by expression → GDEs provide a powerful embedding space for regulatory sequence design.
🦠 SARS-CoV-2 spike protein dynamics
GDEs embed monthly lineage distributions and recover smooth latent chronologies.
GDEs model phenotype distributions induced by perturbations and generalize to unseen conditions.
🧬 DNA methylation (253M reads)
We learn tissue-specific methylation patterns directly from raw bisulfite reads — no alignment necessary.

GDEs model phenotype distributions induced by perturbations and generalize to unseen conditions.
🧬 DNA methylation (253M reads)
We learn tissue-specific methylation patterns directly from raw bisulfite reads — no alignment necessary.
Each clone is a population of cells → a distribution over expression. GDEs predict clonal fate better than prior approaches.
🧪 CRISPR perturbation effects
GDEs can improve zero-shot prediction of transcriptional response distributions.

Each clone is a population of cells → a distribution over expression. GDEs predict clonal fate better than prior approaches.
🧪 CRISPR perturbation effects
GDEs can improve zero-shot prediction of transcriptional response distributions.
We show that GDE embeddings are asymptotically normal, grounding this with results from empirical process theory.

We show that GDE embeddings are asymptotically normal, grounding this with results from empirical process theory.
GDE latent distances track Wasserstein-2 (W₂) distances across modalities (shown for multinomial distributions)
Latent interpolations recover optimal transport paths (shown for Gaussians and Gaussian mixtures)


GDE latent distances track Wasserstein-2 (W₂) distances across modalities (shown for multinomial distributions)
Latent interpolations recover optimal transport paths (shown for Gaussians and Gaussian mixtures)
✅ Abstracts away sampling noise
✅ Enables reasoning at the population level
✅ Supports prediction, comparison, and generation of distributions
✅ Lets us leverages powerful domain-specific generative models to learn distributional structure
✅ Abstracts away sampling noise
✅ Enables reasoning at the population level
✅ Supports prediction, comparison, and generation of distributions
✅ Lets us leverages powerful domain-specific generative models to learn distributional structure
1. an encoder that maps samples to a latent representation and
2. any conditional generative model that samples from the distribution conditional on the latent
to lift autoencoders from points to distributions!
1. an encoder that maps samples to a latent representation and
2. any conditional generative model that samples from the distribution conditional on the latent
to lift autoencoders from points to distributions!
- Cells grouped by clone or patient
- Sequences by tissue, time, or location
And we want to model the clone, the patient, or the tissue, not just the individual points.
- Cells grouped by clone or patient
- Sequences by tissue, time, or location
And we want to model the clone, the patient, or the tissue, not just the individual points.
@bayesianboy.bsky.social, and @zacharylipton.bsky.social! You can find full details in the paper here: arxiv.org/abs/2410.09600.
Looking forward to discussing how we can build more nuanced approaches to fairness in machine learning! 5/5

@bayesianboy.bsky.social, and @zacharylipton.bsky.social! You can find full details in the paper here: arxiv.org/abs/2410.09600.
Looking forward to discussing how we can build more nuanced approaches to fairness in machine learning! 5/5
- Model complex measurement biases
- Encode domain-specific constraints
- Systematically explore metric sensitivity
Stop by our poster today from 4:30-7:30pm in the West Ballroom to learn about rigorous fairness evals and beyond! 4 / 5
- Model complex measurement biases
- Encode domain-specific constraints
- Systematically explore metric sensitivity
Stop by our poster today from 4:30-7:30pm in the West Ballroom to learn about rigorous fairness evals and beyond! 4 / 5


1. academic.oup.com/jrsssb/artic...
2. www.pnas.org/doi/10.1073/...
3. www.pnas.org/doi/abs/10.1...
4. arxiv.org/abs/2303.01422
1. academic.oup.com/jrsssb/artic...
2. www.pnas.org/doi/10.1073/...
3. www.pnas.org/doi/abs/10.1...
4. arxiv.org/abs/2303.01422

