
Mykhaylo M. Malakhov
@mykmal.xyz
Genetic epidemiology postdoc at @stanford.edu
This is an exceedingly late post, but I wanted to share a few pictures from #IGES2025 earlier this fall. I had the honor of giving a platform talk on age-related genetic dysregulation, and I really enjoyed connecting with other researchers and exploring the beautiful city of Cologne!




November 7, 2025 at 11:09 PM
This is an exceedingly late post, but I wanted to share a few pictures from #IGES2025 earlier this fall. I had the honor of giving a platform talk on age-related genetic dysregulation, and I really enjoyed connecting with other researchers and exploring the beautiful city of Cologne!
More votes have been counted since then, and the updated map looks even better. There's not a single red cone across the whole state!

November 7, 2025 at 9:03 AM
More votes have been counted since then, and the updated map looks even better. There's not a single red cone across the whole state!
Yesterday I successfully defended my dissertation! It feels surreal to finally reach the end of my PhD journey at @umnsph.bsky.social. Thank you to my adviser Wei Pan and to my committee members @elock.bsky.social, @aazaidi.bsky.social, and Thierry Chekouo for your invaluable mentorship and support!

July 24, 2025 at 9:24 PM
Yesterday I successfully defended my dissertation! It feels surreal to finally reach the end of my PhD journey at @umnsph.bsky.social. Thank you to my adviser Wei Pan and to my committee members @elock.bsky.social, @aazaidi.bsky.social, and Thierry Chekouo for your invaluable mentorship and support!
I had a great time at STATGEN 2025 last week! Thank you to everyone who came to my talk and to the organizers for putting on this conference.

May 28, 2025 at 7:53 PM
I had a great time at STATGEN 2025 last week! Thank you to everyone who came to my talk and to the organizers for putting on this conference.
Importantly, type I error was not inflated because imputed data was only used for training, not inference. Overall, we demonstrated that GWAS-based trait imputation can be used to increase sample sizes and thereby boost power for nonlinear TWAS/PWAS methods that require individual-level data. (6/6)
May 7, 2025 at 6:12 PM
Importantly, type I error was not inflated because imputed data was only used for training, not inference. Overall, we demonstrated that GWAS-based trait imputation can be used to increase sample sizes and thereby boost power for nonlinear TWAS/PWAS methods that require individual-level data. (6/6)
We found that DeLIVR trained on imputed AD status was able to identify disease-relevant genes that were missed if the models were instead trained on observed data, and the same story held true when identifying putative risk proteins. We also showed that LS-imputation has the best performance. (5/6)

May 7, 2025 at 6:12 PM
We found that DeLIVR trained on imputed AD status was able to identify disease-relevant genes that were missed if the models were instead trained on observed data, and the same story held true when identifying putative risk proteins. We also showed that LS-imputation has the best performance. (5/6)
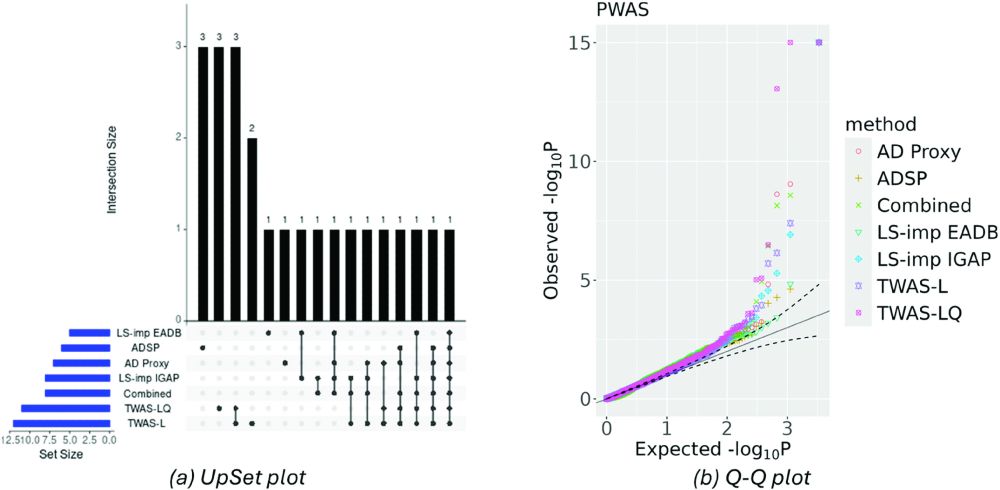
We considered the nonlinear TWAS/PWAS methods DeLIVR (based on deep neural networks) and TWAS-LQ (based on a parametric polynomial model). We focused on Alzheimer's disease (AD) and imputed AD status from family history or from GWAS summary statistics using LS-imputation, PRS-CS, and LDpred2. (4/6)

May 7, 2025 at 6:12 PM
We considered the nonlinear TWAS/PWAS methods DeLIVR (based on deep neural networks) and TWAS-LQ (based on a parametric polynomial model). We focused on Alzheimer's disease (AD) and imputed AD status from family history or from GWAS summary statistics using LS-imputation, PRS-CS, and LDpred2. (4/6)
But linear TWAS/PWAS methods have one major advantage: linear stage 2 models can be estimated using GWAS summary statistics instead of individual-level data. So what if we use GWAS summary statistics to create imputed individual-level datasets and then train nonlinear TWAS/PWAS models on them? (3/6)

May 7, 2025 at 6:12 PM
But linear TWAS/PWAS methods have one major advantage: linear stage 2 models can be estimated using GWAS summary statistics instead of individual-level data. So what if we use GWAS summary statistics to create imputed individual-level datasets and then train nonlinear TWAS/PWAS models on them? (3/6)
I'm excited to share our latest work, now out in #PLOSGenetics! If you feel limited by linear models in TWAS or PWAS but don't think you can switch to nonlinear methods due to a lack of individual-level phenotype data, this paper is for you.
🔗 journals.plos.org/plosgenetics...
Thread below 👇 (1/6)
🔗 journals.plos.org/plosgenetics...
Thread below 👇 (1/6)

May 7, 2025 at 6:12 PM
I'm excited to share our latest work, now out in #PLOSGenetics! If you feel limited by linear models in TWAS or PWAS but don't think you can switch to nonlinear methods due to a lack of individual-level phenotype data, this paper is for you.
🔗 journals.plos.org/plosgenetics...
Thread below 👇 (1/6)
🔗 journals.plos.org/plosgenetics...
Thread below 👇 (1/6)

