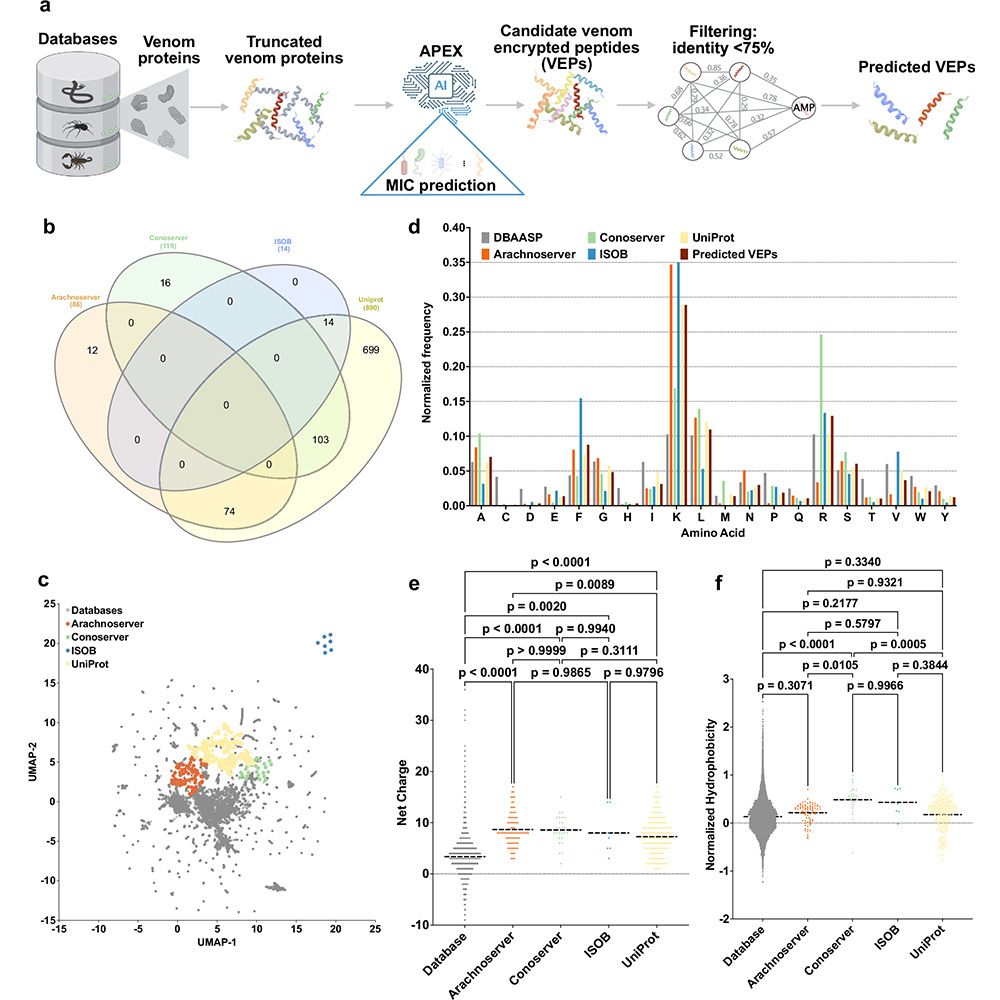

---
#proteomics #prot-preprint

---
#proteomics #prot-preprint
bit.ly/4kEQwqu
#STS

bit.ly/4kEQwqu
#STS
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Key points:
- You don't need gigantic models. The two smaller ESM C variants work great.
- There is huge variability in performance across datasets. We have no idea why.
www.nature.com/articles/s41...
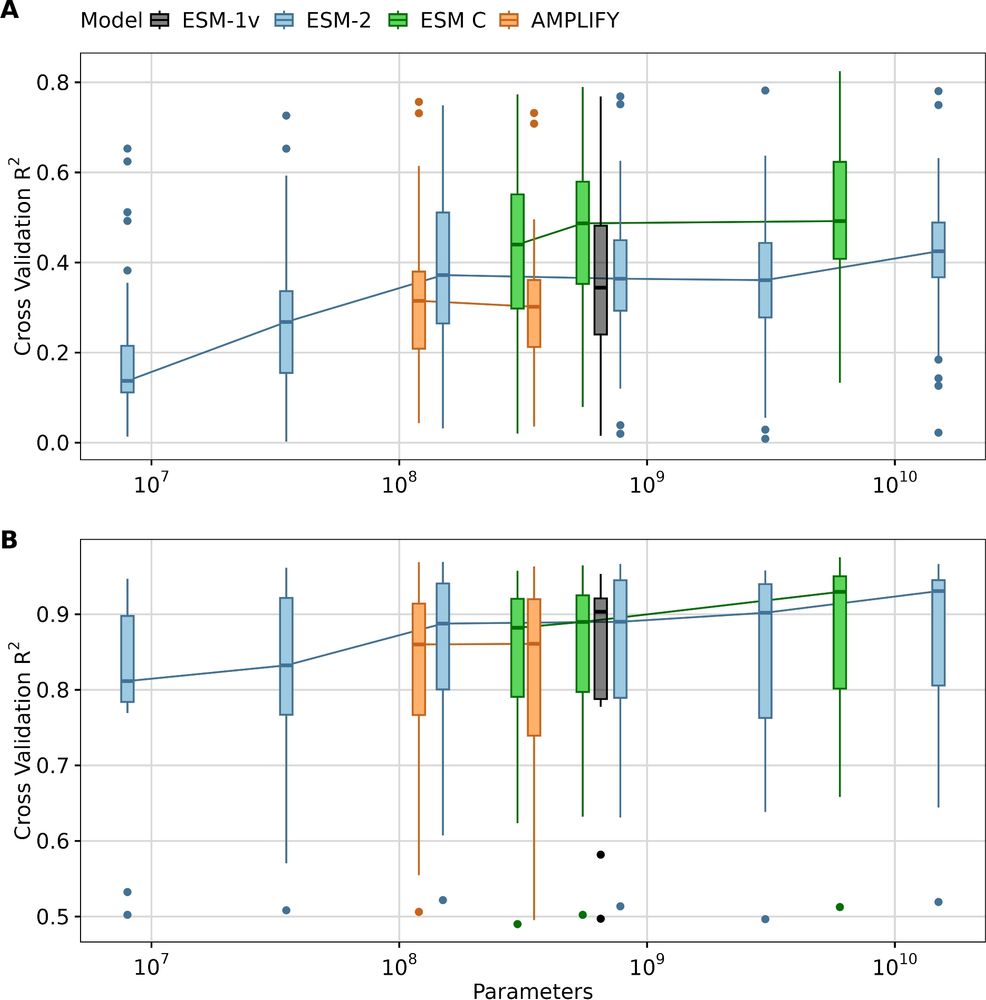

Key points:
- You don't need gigantic models. The two smaller ESM C variants work great.
- There is huge variability in performance across datasets. We have no idea why.
www.nature.com/articles/s41...

