(TLDR; low-affinity motifs matter as pioneers!)

(TLDR; low-affinity motifs matter as pioneers!)


Long-range massively parallel reporter assay reveals rules of distal enhancer-promoter interactions
From Barak Cohen's lab at @washu.bsky.social
Read the pre-print 👇
doi.org/10.1101/2025...
Learn more about the research from the Cohen lab: bclab.wustl.edu
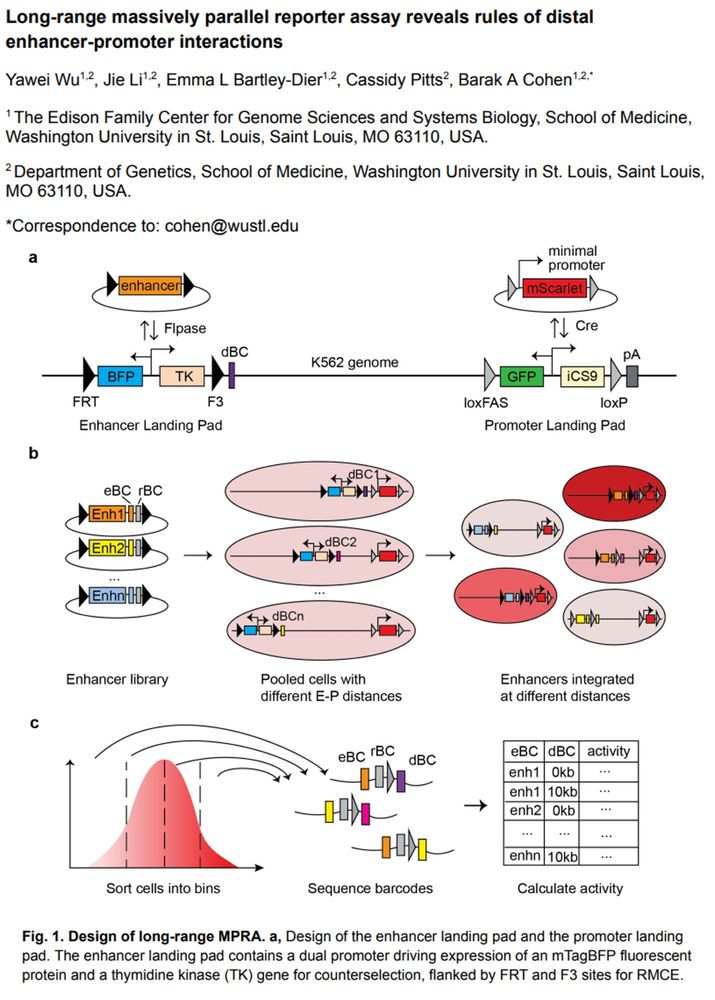
Long-range massively parallel reporter assay reveals rules of distal enhancer-promoter interactions
From Barak Cohen's lab at @washu.bsky.social
Read the pre-print 👇
doi.org/10.1101/2025...
Learn more about the research from the Cohen lab: bclab.wustl.edu
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...

1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...
Read more: bit.ly/41Czvqy

Read more: bit.ly/41Czvqy
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

