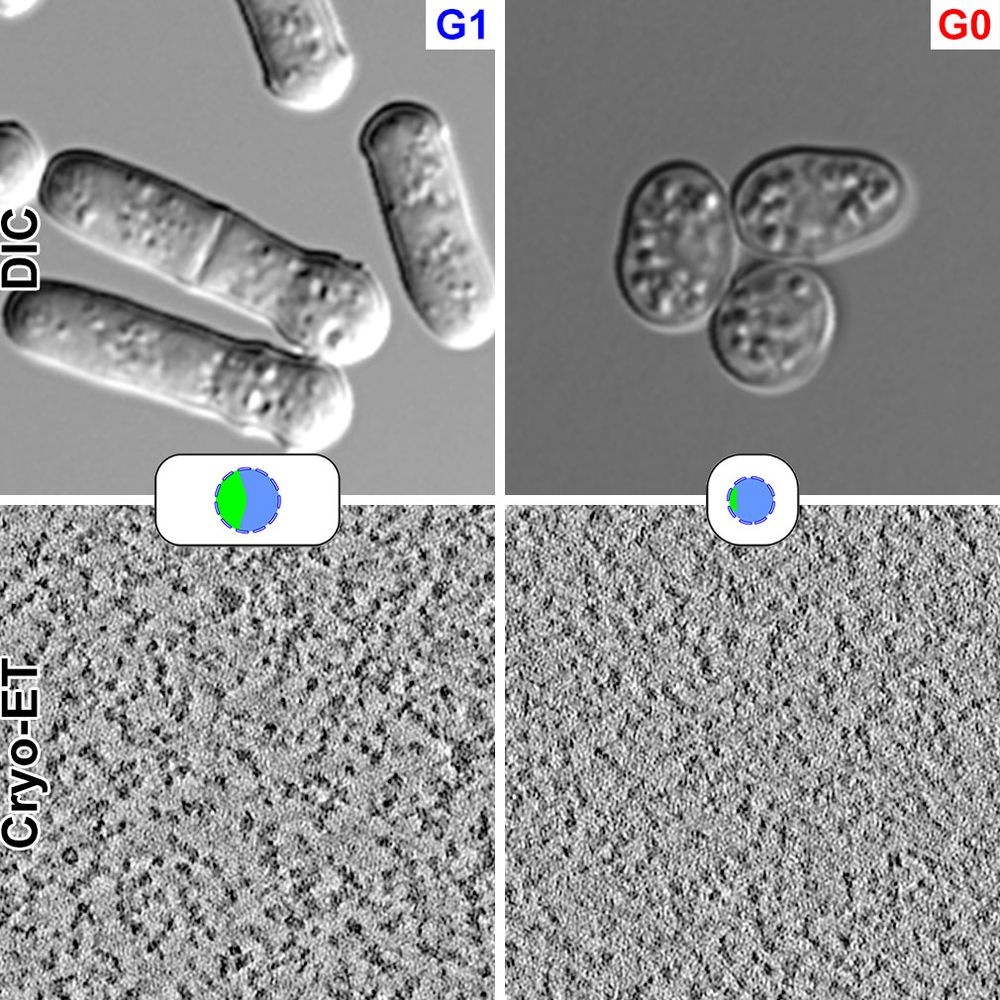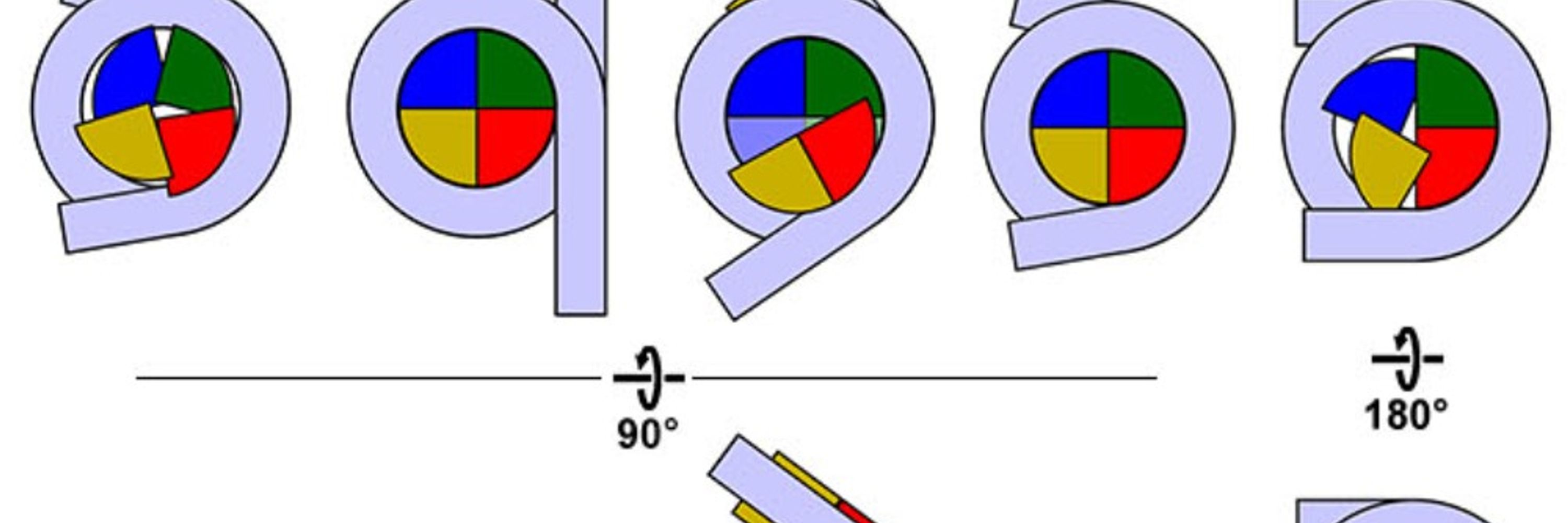
Former @ Dept. of Biological Sciences & CBIS, NUS
anaphase.org
med.virginia.edu/faculty/faculty-listing/wwh6xu
mstdn.science/@MitosisLab



Congrats also to coauthors Tingsheng Liu, @shujuncai.bsky.social, Cai Tong Ng, Jian Shi and collaborators Weimei Ruan Uttam Surana.
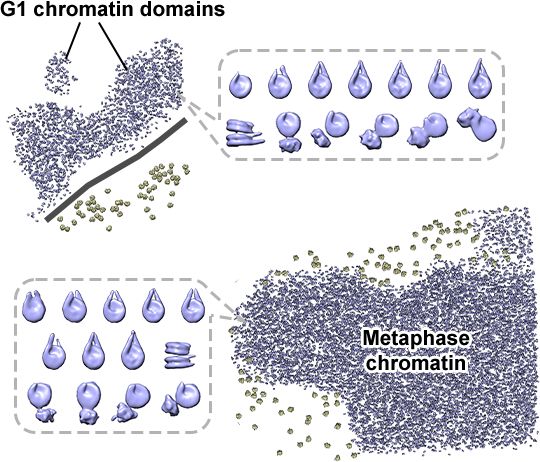
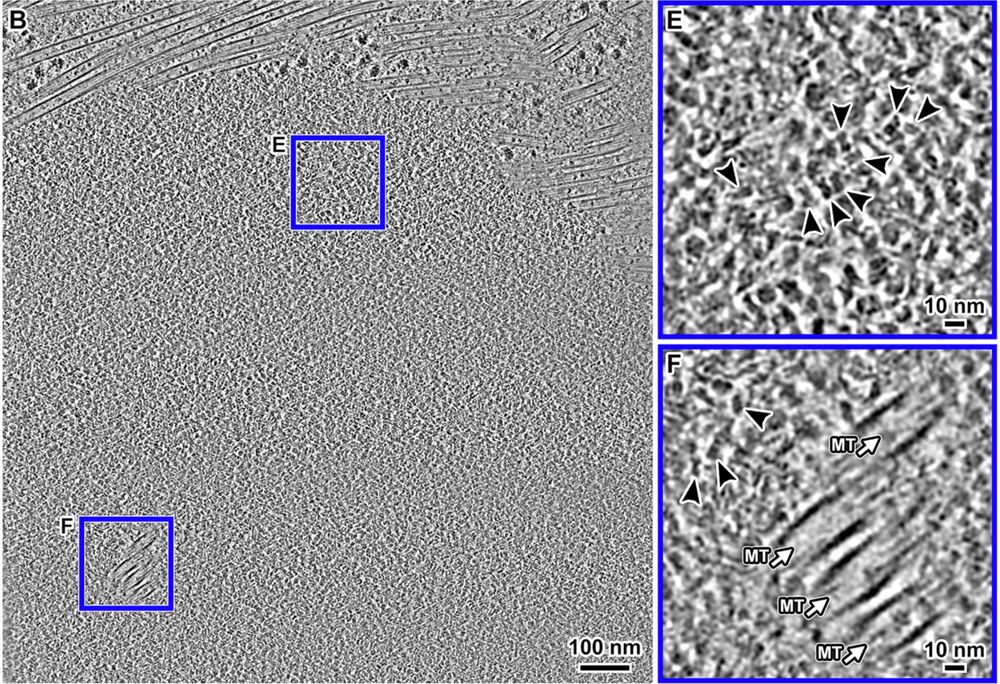
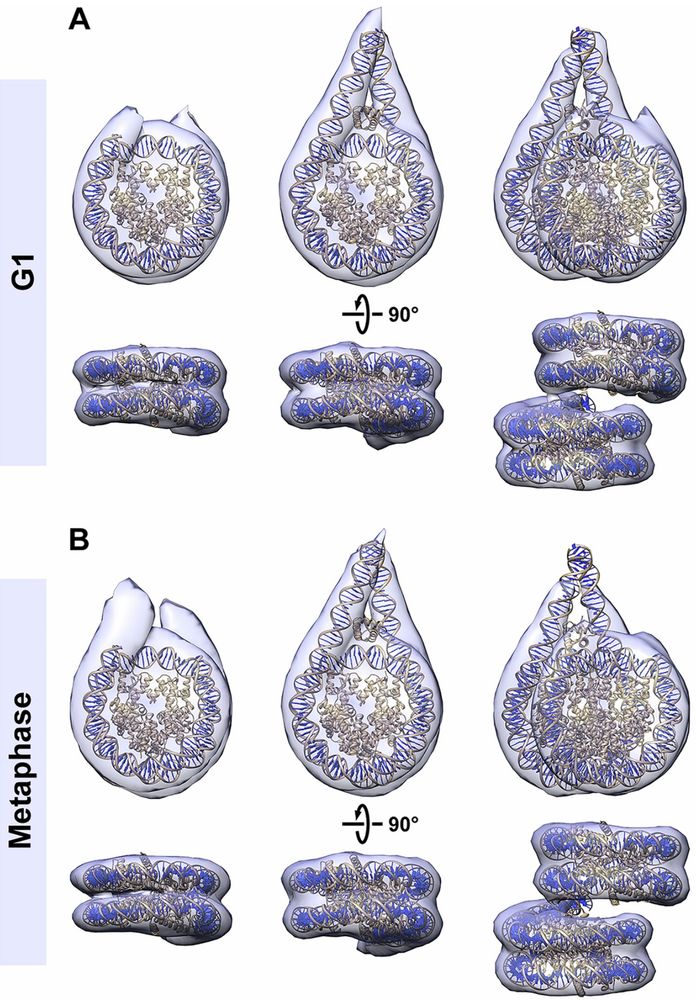
Congrats also to coauthors Tingsheng Liu, @shujuncai.bsky.social, Cai Tong Ng, Jian Shi and collaborators Weimei Ruan Uttam Surana.