Mitch Guttman
@mitchguttman.bsky.social
Molecular biologist interested in non-coding RNAs, nuclear organization, and gene regulation. Professor at Caltech
Pinned
Mitch Guttman
@mitchguttman.bsky.social
· Jul 26

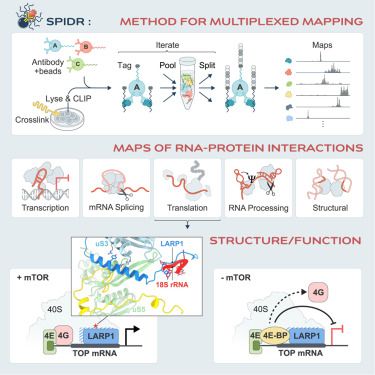
Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @cp-cell.bsky.social, with the Jovanovic lab, we describe SPIDR – a method for mapping the RNA binding sites of dozens of RBPs in a single experiment. www.sciencedirect.com/science/arti...
Reposted by Mitch Guttman
SPIDR enables multiplexed mapping of RNA-protein interactions and uncovers a mechanism for selective translational suppression upon cell stress @cellcellpress.bsky.social @mitchguttman.bsky.social
www.cell.com/cell/fulltex...
www.cell.com/cell/fulltex...
July 22, 2025 at 6:23 PM
SPIDR enables multiplexed mapping of RNA-protein interactions and uncovers a mechanism for selective translational suppression upon cell stress @cellcellpress.bsky.social @mitchguttman.bsky.social
www.cell.com/cell/fulltex...
www.cell.com/cell/fulltex...
Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @cp-cell.bsky.social, with the Jovanovic lab, we describe SPIDR – a method for mapping the RNA binding sites of dozens of RBPs in a single experiment. www.sciencedirect.com/science/arti...

July 26, 2025 at 7:15 PM
Many proteins bind RNA, yet we still don’t know what RNAs most bind because methods map one RBP at a time. In @cp-cell.bsky.social, with the Jovanovic lab, we describe SPIDR – a method for mapping the RNA binding sites of dozens of RBPs in a single experiment. www.sciencedirect.com/science/arti...
Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
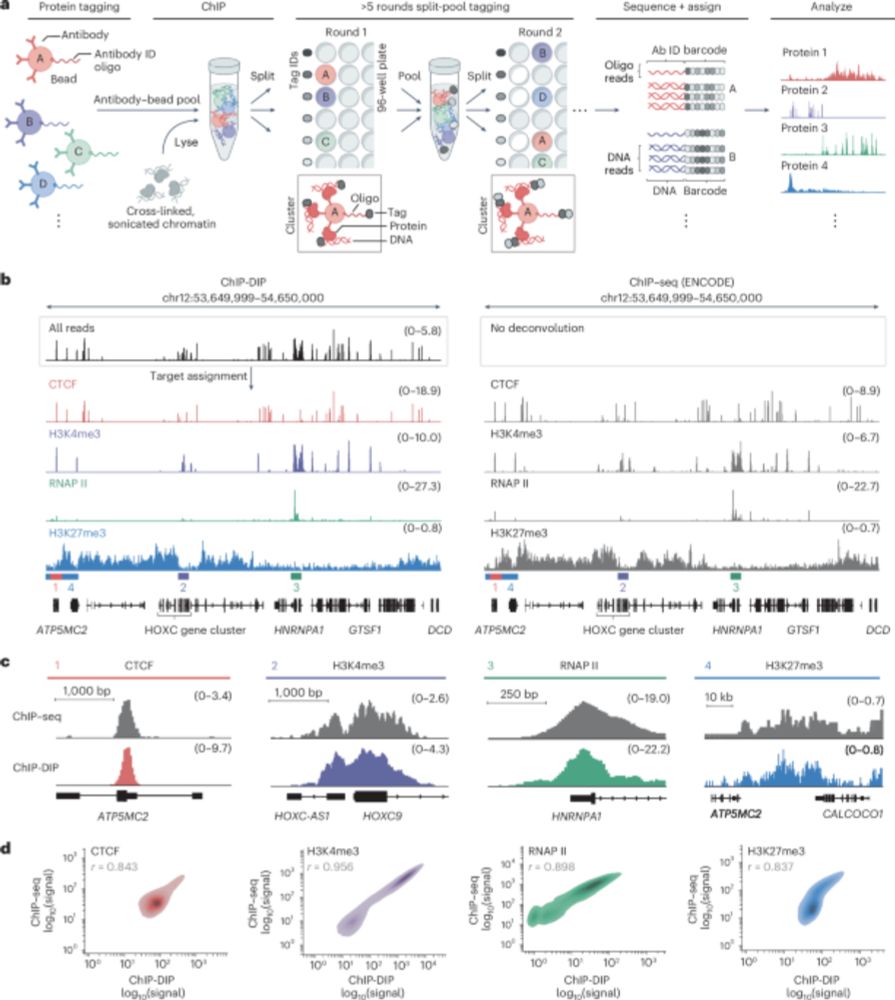
ChIP-DIP maps binding of hundreds of proteins to DNA simultaneously and identifies diverse gene regulatory elements - Nature Genetics
ChIP-DIP (ChIP done in parallel) is a highly multiplex assay for protein–DNA binding, scalable to hundreds of proteins including modified histones, chromatin regulators and transcription factors, offe...
www.nature.com
November 27, 2024 at 4:13 AM
Gene regulation involves thousands of proteins that bind DNA, yet comprehensively mapping these is challenging. Our paper in Nature Genetics describes ChIP-DIP, a method for genome-wide mapping of hundreds of DNA-protein interactions in a single experiment.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Mitch Guttman
Great thread on our paper from Jan👇
Our latest paper, a fantastic collaboration with the Mendell Lab, is out. Great work by Xiaoqiang Zhu and Victor Cruz. We uncover how tRNA structural features in the ribosomal P-site modulate co-translational mRNA decay by the CCR4-NOT complex. #RNAsky #RNAbiology #translation #ribosome #cryoEM 1/5
www.science.org
November 22, 2024 at 6:39 PM
Great thread on our paper from Jan👇
A report made counterintuitive claims that PRC2 binds
→ more RNA than PTBP1 & hnRNPU
→ RNAs that do not exist in the cell
We found a simple explanation: the authors ignore most RNA in the sample, leading to inflated background & misleading conclusions.
www.biorxiv.org/content/10.1...
→ more RNA than PTBP1 & hnRNPU
→ RNAs that do not exist in the cell
We found a simple explanation: the authors ignore most RNA in the sample, leading to inflated background & misleading conclusions.
www.biorxiv.org/content/10.1...

Failing to account for RNA quantity inflates background and leads to the misleading appearance that PRC2 and GFP bind to RNA in vivo
We recently published biochemical and quantitative evidence that challenges the widespread claims that PRC2 binds directly to many RNAs in vivo . A recent preprint performs a re-analysis of some of ou...
www.biorxiv.org
November 18, 2024 at 1:11 AM
A report made counterintuitive claims that PRC2 binds
→ more RNA than PTBP1 & hnRNPU
→ RNAs that do not exist in the cell
We found a simple explanation: the authors ignore most RNA in the sample, leading to inflated background & misleading conclusions.
www.biorxiv.org/content/10.1...
→ more RNA than PTBP1 & hnRNPU
→ RNAs that do not exist in the cell
We found a simple explanation: the authors ignore most RNA in the sample, leading to inflated background & misleading conclusions.
www.biorxiv.org/content/10.1...
Reposted by Mitch Guttman
Leonid Mirny and I wrote this for all interested in chromosomes: "The chromosome folding problem and how cells solve it"
www.cell.com/action/showP...
www.cell.com/action/showP...
www.cell.com
November 14, 2024 at 4:02 PM
Leonid Mirny and I wrote this for all interested in chromosomes: "The chromosome folding problem and how cells solve it"
www.cell.com/action/showP...
www.cell.com/action/showP...

