
Inhibition of the key regulator of non-canonical NF-kB (NIK) totally neutralises the drug resistance caused by the TME in DLBCL🎉
5/🧵

Inhibition of the key regulator of non-canonical NF-kB (NIK) totally neutralises the drug resistance caused by the TME in DLBCL🎉
5/🧵
Good news: this means Ibrutinib overcomes the TME!
4/🧵

Good news: this means Ibrutinib overcomes the TME!
4/🧵
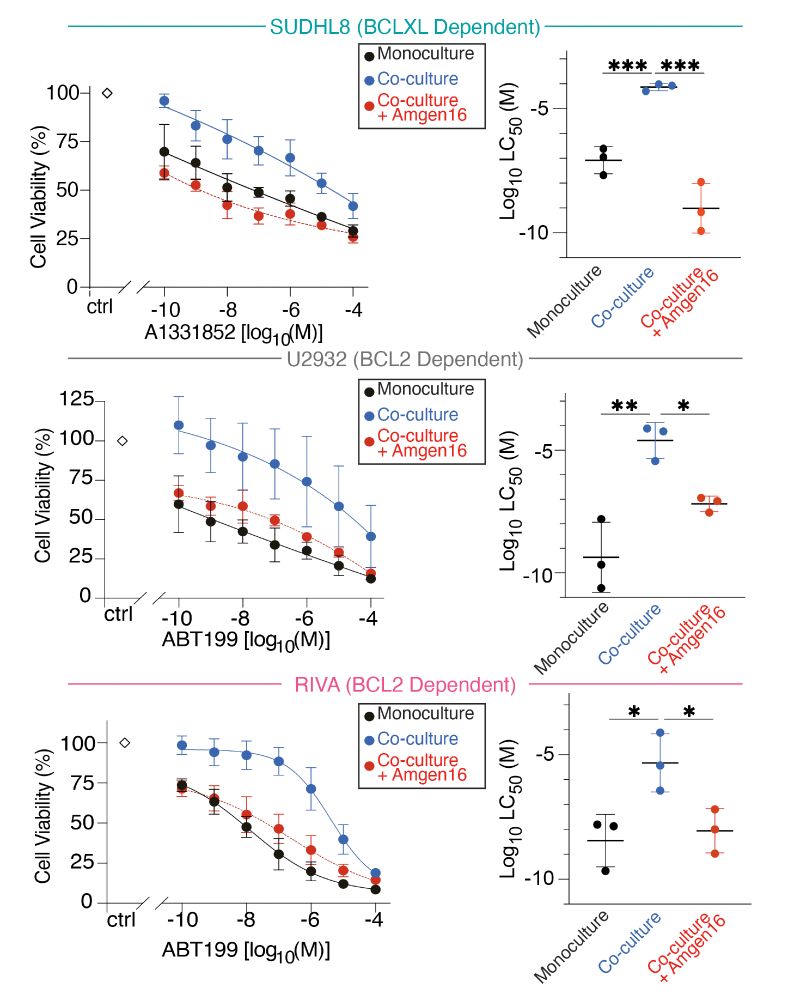
- culturing DLBCL cells with cells expressing CD40 (to mimick the TME) creates resistance to BH3-mimetics.
- this happens whether you're targeting BCL2 or BCLXL.
- Compensatory upregulation of BCLXL causes resistance.
- In SUDHL8 cells MCL1 also increases. Why!?
3/🧵

- culturing DLBCL cells with cells expressing CD40 (to mimick the TME) creates resistance to BH3-mimetics.
- this happens whether you're targeting BCL2 or BCLXL.
- Compensatory upregulation of BCLXL causes resistance.
- In SUDHL8 cells MCL1 also increases. Why!?
3/🧵
- Established NFκB -> BCL2 link is actually a bit fuzzy (which NFκB subunits control which BCL2 proteins in DLBCL?)
- scRNAseq suggests tumour microenvironment (TME) activates non-canonical NFκB (RelB:p52).
- Can we overcome the protective effect of the TME ?
2/🧵

- Established NFκB -> BCL2 link is actually a bit fuzzy (which NFκB subunits control which BCL2 proteins in DLBCL?)
- scRNAseq suggests tumour microenvironment (TME) activates non-canonical NFκB (RelB:p52).
- Can we overcome the protective effect of the TME ?
2/🧵
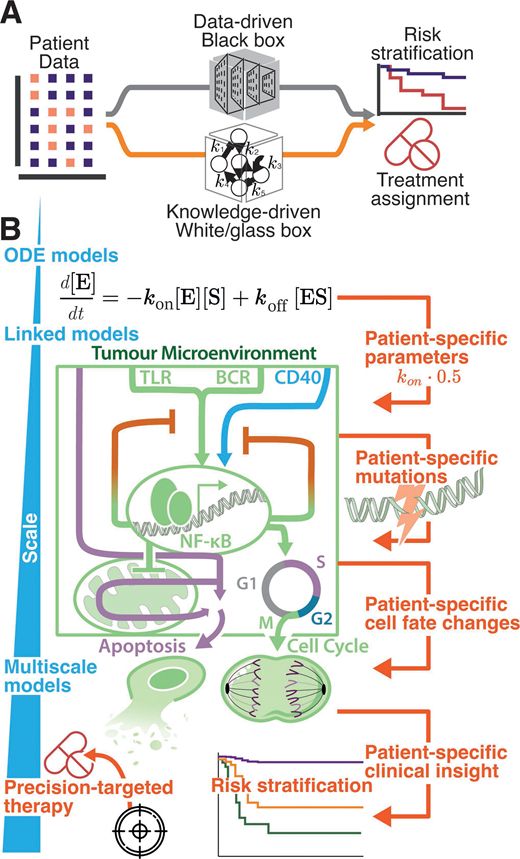
We review:
- how computational biology is helping us understand lymphoma,
- how machine learning and cell simulations make sense of vast amounts of data,
- how models can personalise treatments to each person with lymphoma.
portlandpress.com/biochemsoctr...
We review:
- how computational biology is helping us understand lymphoma,
- how machine learning and cell simulations make sense of vast amounts of data,
- how models can personalise treatments to each person with lymphoma.
portlandpress.com/biochemsoctr...
Great venue and even better science.

Great venue and even better science.
It's so rewarding to see our lab's first PhD student graduate, and I couldn't have asked for a better first student.
Job well done Arran!
🤜🤛

It's so rewarding to see our lab's first PhD student graduate, and I couldn't have asked for a better first student.
Job well done Arran!
🤜🤛
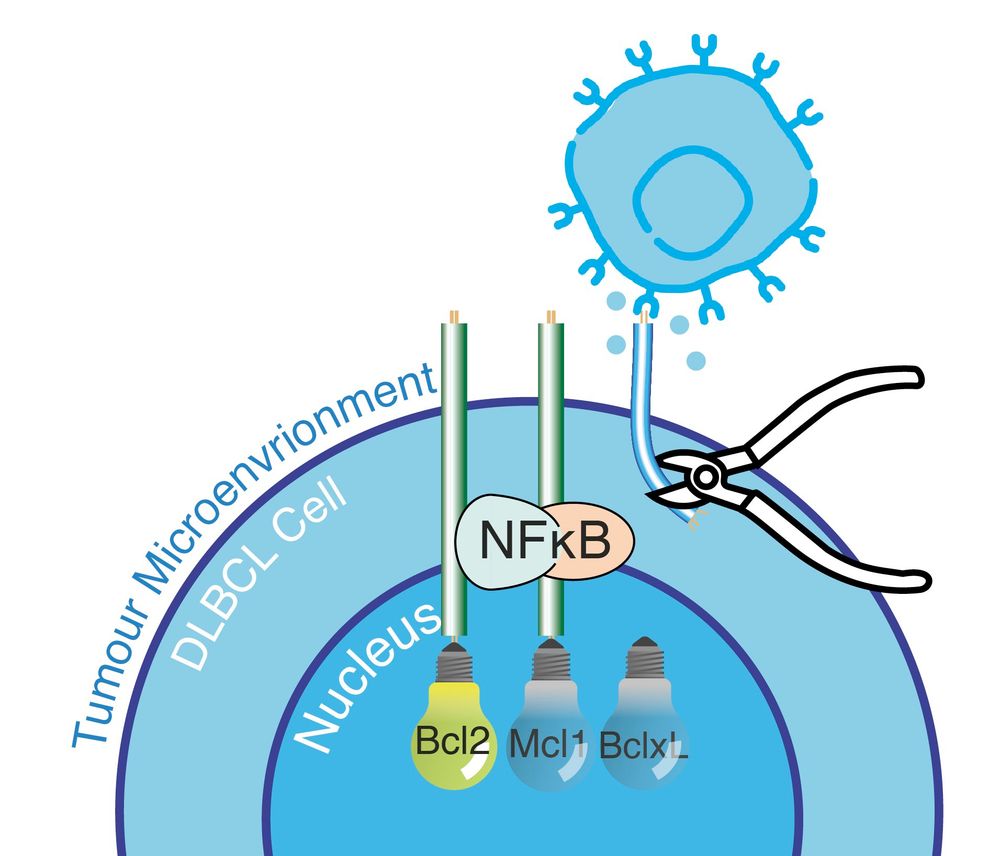
Non-canonical NF-kB inhibition with Amgen16 overcomes all the microenvrionment mediated resistance we discovered, regardless of the mechanism!
@amgen.bsky.social
Let's summarise...
Non-canonical NF-kB inhibition with Amgen16 overcomes all the microenvrionment mediated resistance we discovered, regardless of the mechanism!
@amgen.bsky.social
Let's summarise...
- Different Achilles heel in each DLBCL
- Different resistance mechanism mediated by complicated cross talk.
This makes it really hard to improve treatments.
We asked if we could break all the links with the TME by inhibiting non-canonical NF-kB?
- Different Achilles heel in each DLBCL
- Different resistance mechanism mediated by complicated cross talk.
This makes it really hard to improve treatments.
We asked if we could break all the links with the TME by inhibiting non-canonical NF-kB?
The model told us an IkBe knockout would selectively upregulate cRel, which would upregulate MCL1 if we were right.
Collaborating with Alexander Hoffmann's group we tested this in IkBe KO mice.
cRel and MCL1 go up!
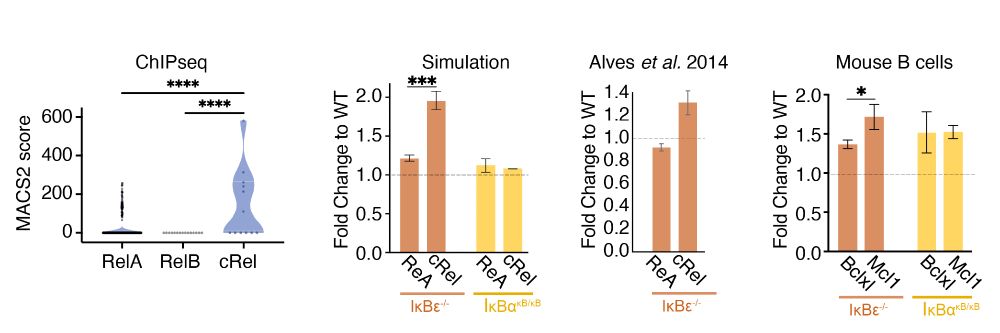
The model told us an IkBe knockout would selectively upregulate cRel, which would upregulate MCL1 if we were right.
Collaborating with Alexander Hoffmann's group we tested this in IkBe KO mice.
cRel and MCL1 go up!
Chronic NF-kB activation creates crosstalk between the pathways.
We also knew this chronic NF-kB was causing the drug resistance, because when we inhibit it (with ibrutinib) we can re-sensitize the cells.
But the link between cRel and MCL1 is new.
Is it real?
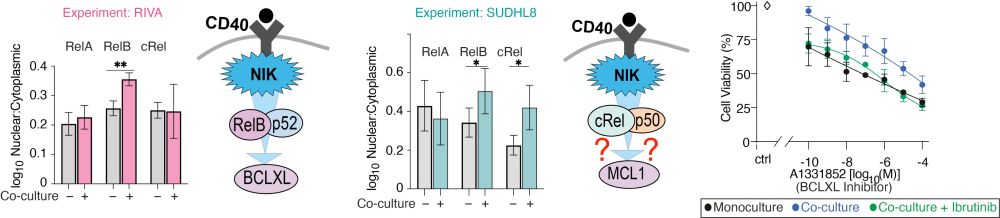
Chronic NF-kB activation creates crosstalk between the pathways.
We also knew this chronic NF-kB was causing the drug resistance, because when we inhibit it (with ibrutinib) we can re-sensitize the cells.
But the link between cRel and MCL1 is new.
Is it real?
A model developed by Arran Pack
predicted that the high RelA could create cross talk between the NF-kB pathways.
CD40 -> NIK -> cRel -> MCL1?
Soumen Basak has seen this before in healthy B-cells. We predict it's happening in DLBCL.
Let's check
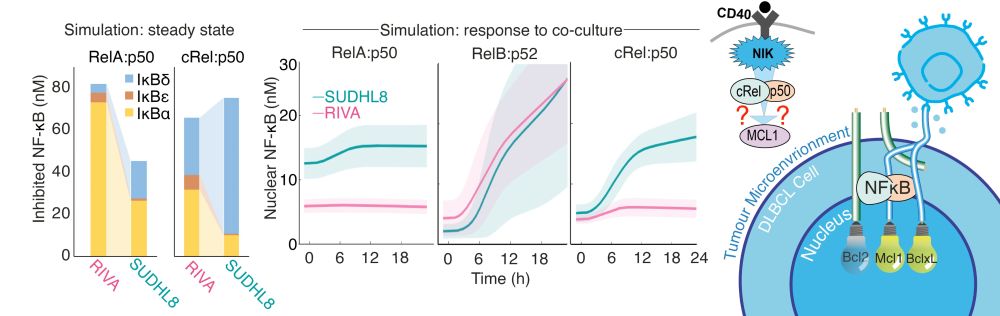
A model developed by Arran Pack
predicted that the high RelA could create cross talk between the NF-kB pathways.
CD40 -> NIK -> cRel -> MCL1?
Soumen Basak has seen this before in healthy B-cells. We predict it's happening in DLBCL.
Let's check
We used imaging to find which NF-kB components were in the nucleus before the cells went into co-culture.
Lots of RelA in cells that induce MCL1.
But how does high RelA in monoculture lead to MCL1 in co-culture?

We used imaging to find which NF-kB components were in the nucleus before the cells went into co-culture.
Lots of RelA in cells that induce MCL1.
But how does high RelA in monoculture lead to MCL1 in co-culture?
Some of this we understood:
CD40 -> NIK -> RelB -> BCLXL
But some really did not:
CD40 -> NIK -> ? -> MCL!?
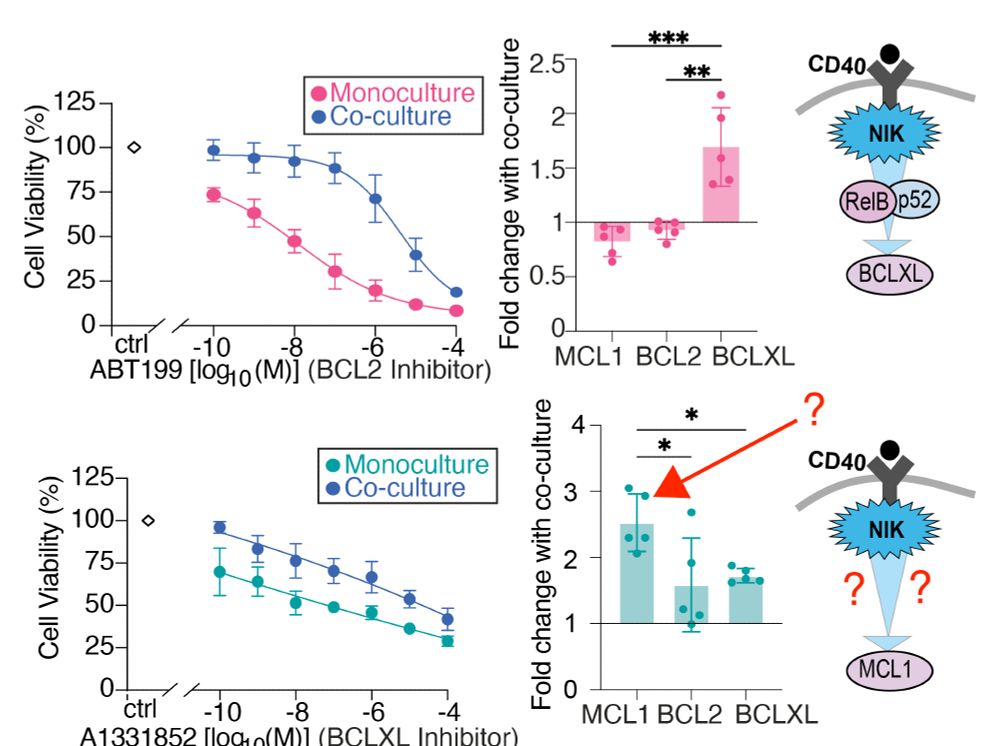
Some of this we understood:
CD40 -> NIK -> RelB -> BCLXL
But some really did not:
CD40 -> NIK -> ? -> MCL!?
This made both cell populations resist treatment. They upregulated BCLXL which compensates for BCL2 inhibition.
Inherent resistance, and micro-environmental resistance in one cell line.
Neat
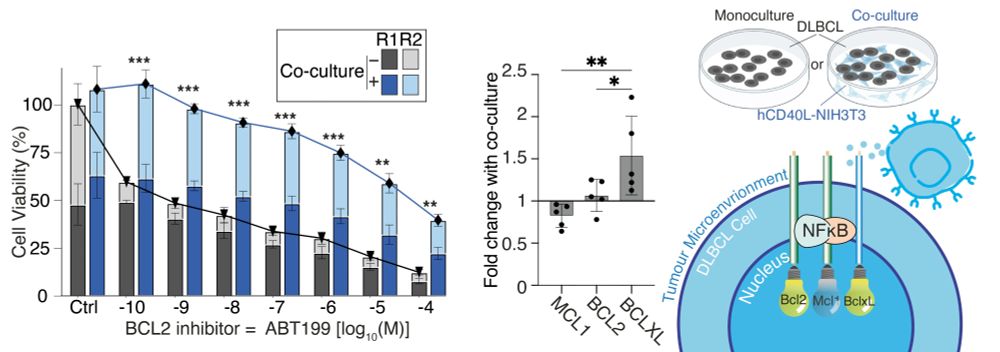
This made both cell populations resist treatment. They upregulated BCLXL which compensates for BCL2 inhibition.
Inherent resistance, and micro-environmental resistance in one cell line.
Neat
Why?
One has high MCL1 to compensate for BCL2 inhibition.
This is cool, but these are cells in a dish. Could we still drug these in a supportive lymph node?
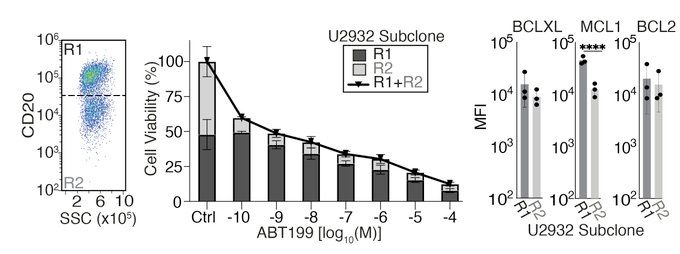
Why?
One has high MCL1 to compensate for BCL2 inhibition.
This is cool, but these are cells in a dish. Could we still drug these in a supportive lymph node?
Yes!
But what is going on with the U2932 cell line? Like lots of cancer it contains multiple cell populations with different genetics.
This makes it great for figuring out how different cells in the same tumour might respond to a drug.
Yes!
But what is going on with the U2932 cell line? Like lots of cancer it contains multiple cell populations with different genetics.
This makes it great for figuring out how different cells in the same tumour might respond to a drug.
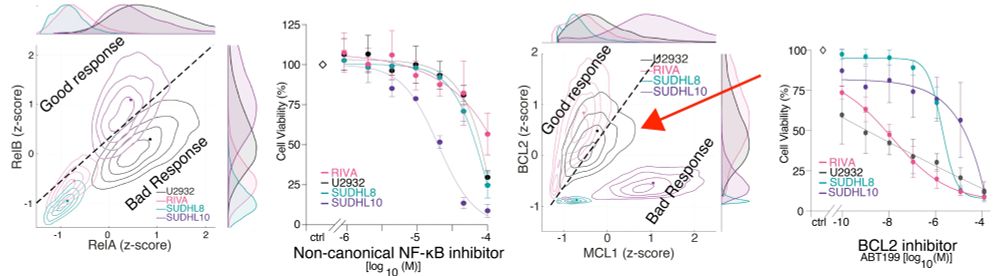
Lots of people have tried inhibiting these. But NF-kB is 2 pathways, 5 proteins, and there are many BCL2-family proteins.
We found previously (doi.org/10.3389/fonc...), each DLBCL has a unique "fingerprint".

Lots of people have tried inhibiting these. But NF-kB is 2 pathways, 5 proteins, and there are many BCL2-family proteins.
We found previously (doi.org/10.3389/fonc...), each DLBCL has a unique "fingerprint".
biorxiv.org/content/10.1...
She asked why drugs that kill cancer cells in blood don't kill them in lymph nodes? How we can fix that?
🧵🧪
@sussexcancer.org
@bsmsmedschool.bsky.social @sussexuni.bsky.social

biorxiv.org/content/10.1...
She asked why drugs that kill cancer cells in blood don't kill them in lymph nodes? How we can fix that?
🧵🧪
@sussexcancer.org
@bsmsmedschool.bsky.social @sussexuni.bsky.social

