#newlab in SR-TIGET June 2025
| #Chromatin | #Microscopy | #FirstGen
www.gabrielelab.com


back.fondazionetelethon.it/uploads/2025...
back.fondazionetelethon.it/uploads/2025...
We ran simulations to estimate EP search times based on 1) measured subdiffusion, 2) physical/genomic distance. Within 300nm (~<200 kb), a gene can find 1000s of enhancers in minutes. Such encounters can’t be selective → EP selectivity needs other mechanisms, e.g. biochemical
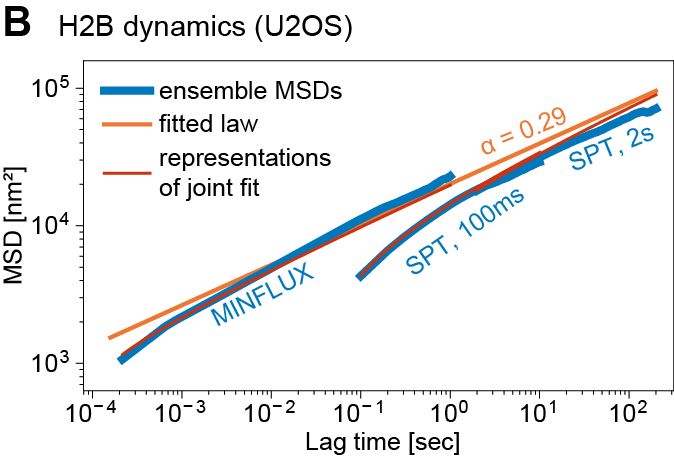
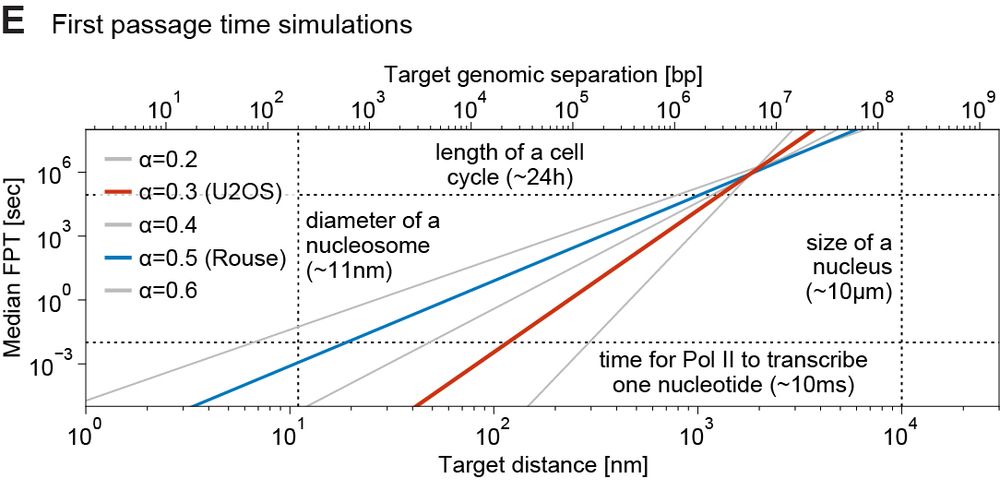
We ran simulations to estimate EP search times based on 1) measured subdiffusion, 2) physical/genomic distance. Within 300nm (~<200 kb), a gene can find 1000s of enhancers in minutes. Such encounters can’t be selective → EP selectivity needs other mechanisms, e.g. biochemical
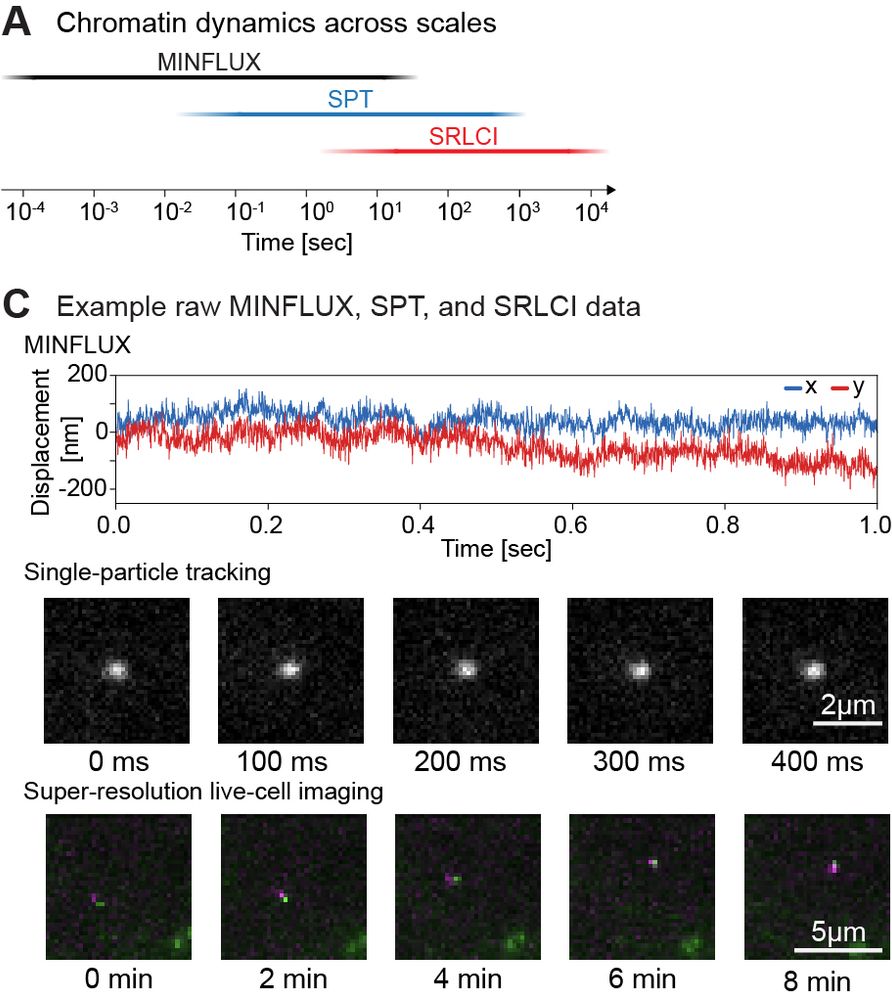
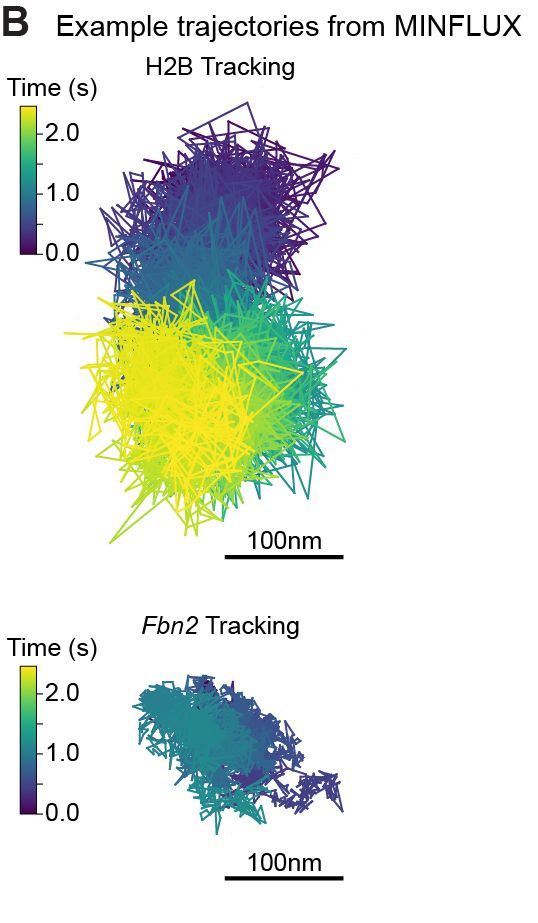
Super excited to share my first preprint from @andersshansen.bsky.social Lab! We used MINFLUX to track chromatin (H2B-Halo and Fbn2 locus) at an unprecedented 200 μs, then combined it with SPT to span μs-minutes (H2B) or SPT & Super-Res Live-Cell Imaging (SRLCI) to span μs-hours (Fbn2)
I am very excited to share that starting this summer, I will open my laboratory at the San Raffaele Telethon Institute for Gene Therapy (SR-Tiget) in Milan, Italy! www.linkedin.com/feed/update/... #newlab #chromatin #epigenetics #milan #chromatindynamics www.gabrielelab.com

I am very excited to share that starting this summer, I will open my laboratory at the San Raffaele Telethon Institute for Gene Therapy (SR-Tiget) in Milan, Italy! www.linkedin.com/feed/update/... #newlab #chromatin #epigenetics #milan #chromatindynamics www.gabrielelab.com
Full paper here:
www.biorxiv.org/content/10.1...
Thread below 🧵
Full paper here:
www.biorxiv.org/content/10.1...
Thread below 🧵

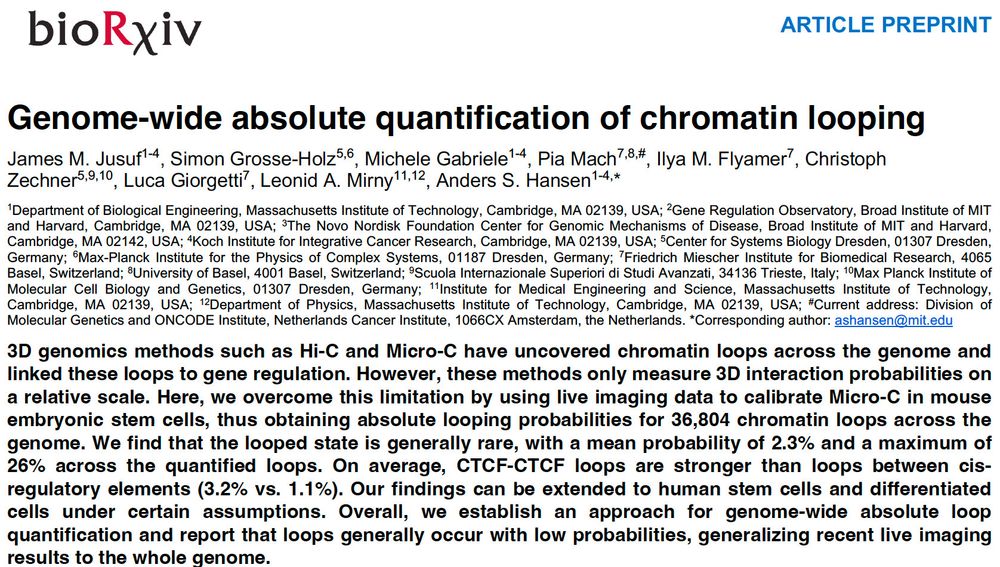
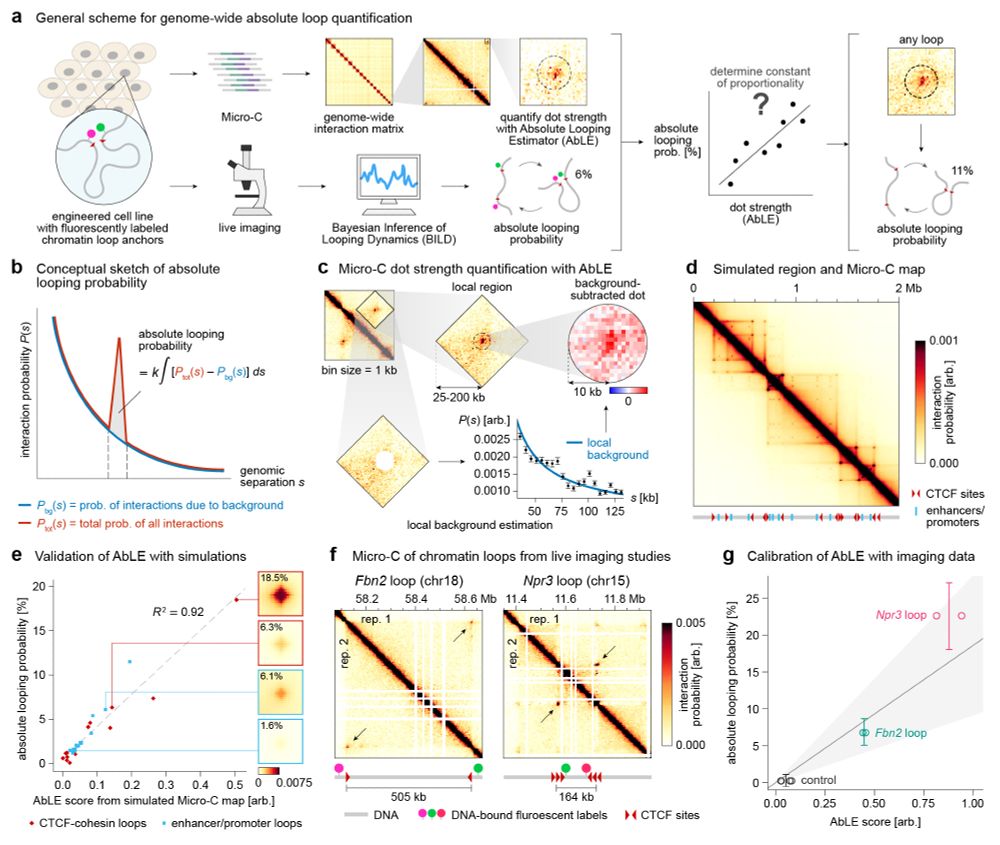
By integrating Micro-C with SuperRes Live-Imaging we can calibrate genomics&imaging to perform absolute quantification of looping (e.g. this loop is present 3%)
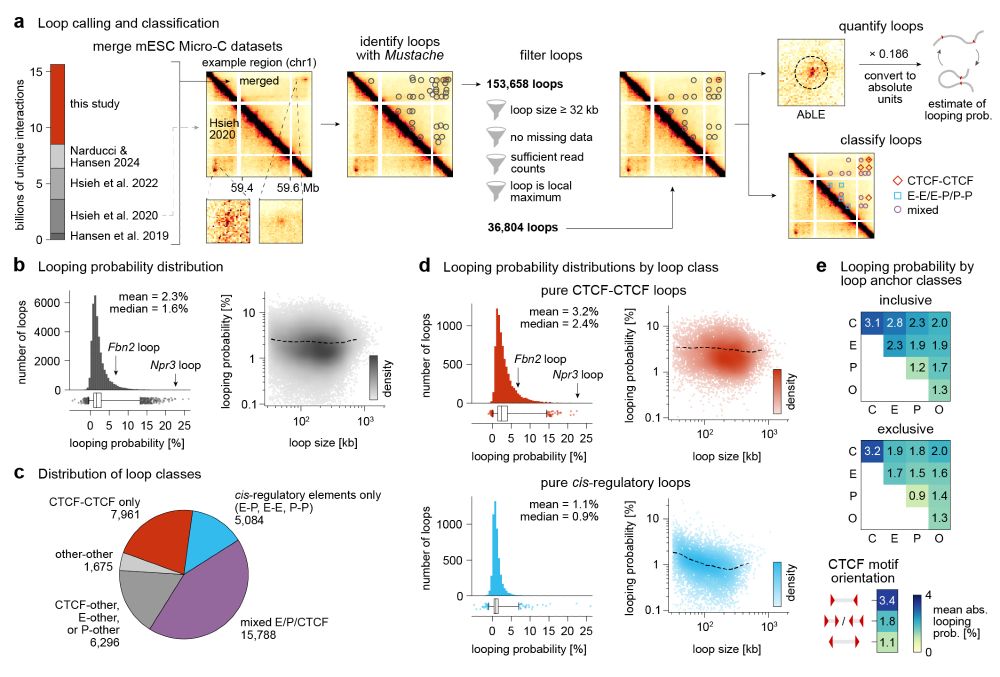
We quantify mESC 36k loops:
www.biorxiv.org/content/10.1...