
@ml4proteins.bsky.social co-organizer
@unc-bcbp.bsky.social for organizing! Gonna be an exciting time!
cbs.web.unc.edu

@unc-bcbp.bsky.social for organizing! Gonna be an exciting time!
cbs.web.unc.edu
Our first valentines 2 years ago, went to breakfast then on a tour of an HTC cluster!! It was this biochemist’s first time seeing a server room😅 then legos. Hate being in grad school at 2 different places now, but still love you!




Our first valentines 2 years ago, went to breakfast then on a tour of an HTC cluster!! It was this biochemist’s first time seeing a server room😅 then legos. Hate being in grad school at 2 different places now, but still love you!





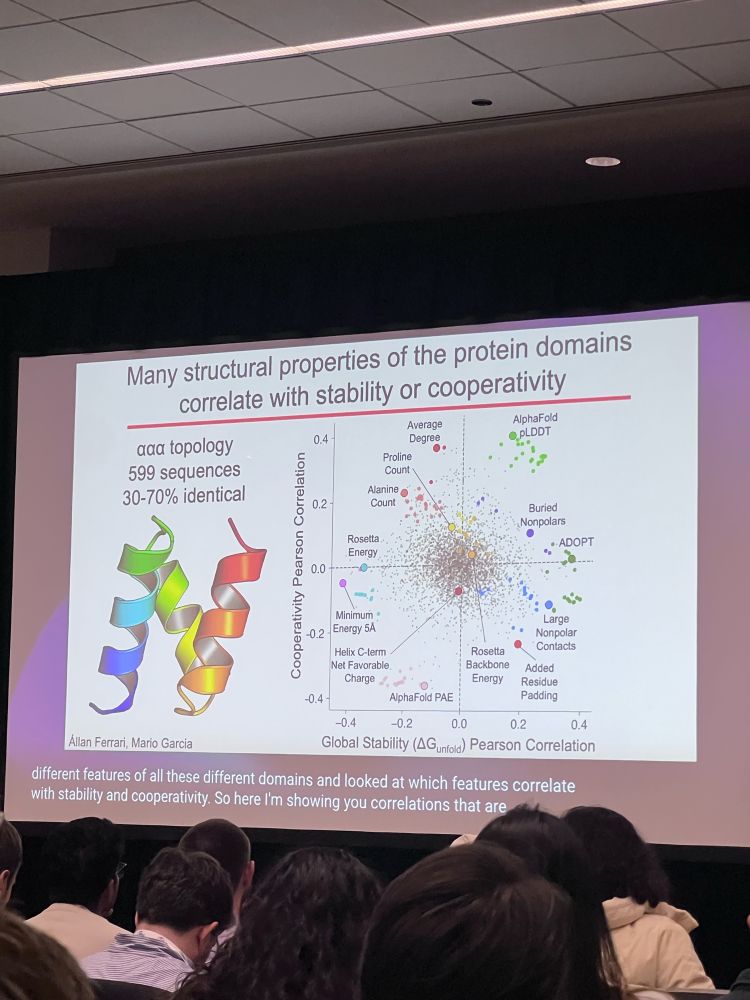
•supported w/ natural domains sharing similar global stability but diff conformational dynamics; to be more open & “less cooperative”, likely indicates hydrophobic chains


•supported w/ natural domains sharing similar global stability but diff conformational dynamics; to be more open & “less cooperative”, likely indicates hydrophobic chains
•coupling H/DX with mass spec, can capture ideas about stability and protein domains and their “opening energy” profile; tells you about partially unfolded status but not specifically where at in the protein

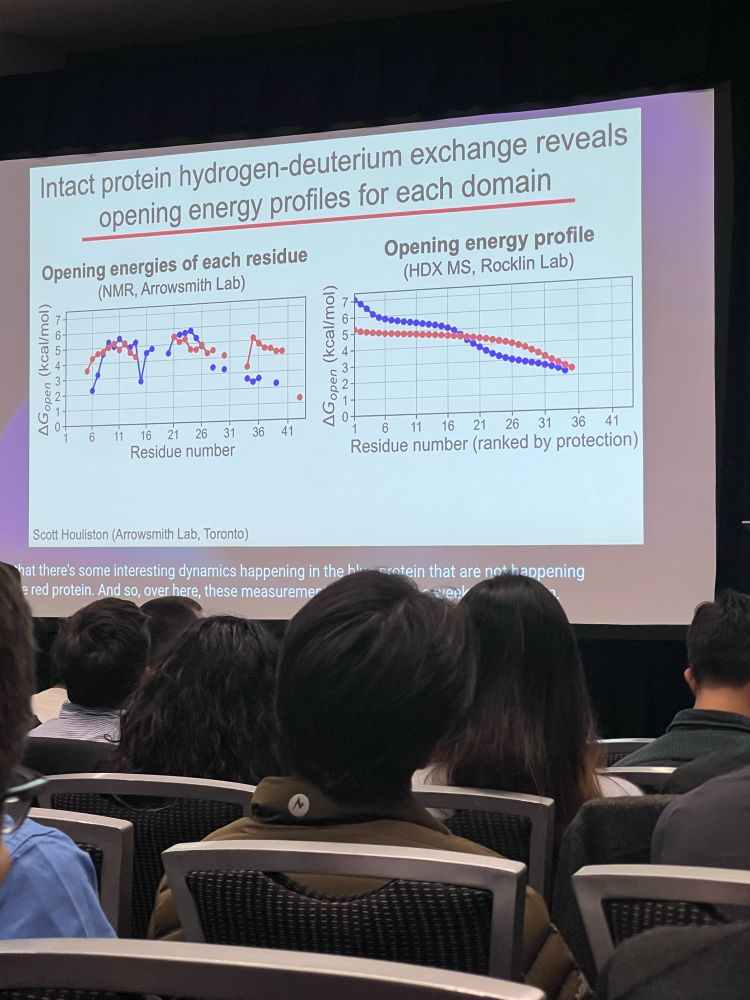
•coupling H/DX with mass spec, can capture ideas about stability and protein domains and their “opening energy” profile; tells you about partially unfolded status but not specifically where at in the protein
•important to understand b/c partial folding/unfolding can have biological relevance to function


•important to understand b/c partial folding/unfolding can have biological relevance to function
•evo-tuning on high likelihood wild types harmed performance
•thus model fitness is truly a matter of preference
•much more also to dive into with their paper


•evo-tuning on high likelihood wild types harmed performance
•thus model fitness is truly a matter of preference
•much more also to dive into with their paper

•also, token distributions are taught in training


•also, token distributions are taught in training
•commented on biases in evolutionary signals from Tree of life used to train pLMs (a favorite paper I read in 2024: shorturl.at/fbC7g)

•commented on biases in evolutionary signals from Tree of life used to train pLMs (a favorite paper I read in 2024: shorturl.at/fbC7g)

•Non-sequence adjacent learning of active site side chains
•also saw good hydrophobicity trends w transmembranes
•lots more to unpack in both the PLAID and CHEAP papers

•Non-sequence adjacent learning of active site side chains
•also saw good hydrophobicity trends w transmembranes
•lots more to unpack in both the PLAID and CHEAP papers
•then compared diversity vs quality across protein length (64-512)
• compared to other models, PLAID showed less degradation in its performance w/ increased protein length

•then compared diversity vs quality across protein length (64-512)
• compared to other models, PLAID showed less degradation in its performance w/ increased protein length
•but, ESMFold latent space represents a joint embedding of sequence structure
•thus can train a latent diffusion objective on the joint embedding space, which is compressed by CHEAP encoder (shorturl.at/fshN4)

•but, ESMFold latent space represents a joint embedding of sequence structure
•thus can train a latent diffusion objective on the joint embedding space, which is compressed by CHEAP encoder (shorturl.at/fshN4)
•structure-based methods are often backbone only
• Sequence-based methods are required to fill in side chains
•all atoms methods go between struc preds and inverse folding
•often not multimodal, don’t jointly sample…


•structure-based methods are often backbone only
• Sequence-based methods are required to fill in side chains
•all atoms methods go between struc preds and inverse folding
•often not multimodal, don’t jointly sample…


•types of questions it is or could probe in the future




•types of questions it is or could probe in the future
•introduced GROQ-seq and utility of HT-experiments
•data quality across multiple locations




•introduced GROQ-seq and utility of HT-experiments
•data quality across multiple locations
•some in field ask what will be the “next AF moment in biology”
•pivoted that question and motivated towards thinking abt a new way to ask about biological data collection process to solve new problems in biology



•some in field ask what will be the “next AF moment in biology”
•pivoted that question and motivated towards thinking abt a new way to ask about biological data collection process to solve new problems in biology
•she also switched gears from mathematical sims in astrophysics to biology and later on proteins ;)
•mission: “interface of biology, automation, and machine learning to rapidly convert the life sciences to a data-first discipline”


•she also switched gears from mathematical sims in astrophysics to biology and later on proteins ;)
•mission: “interface of biology, automation, and machine learning to rapidly convert the life sciences to a data-first discipline”


