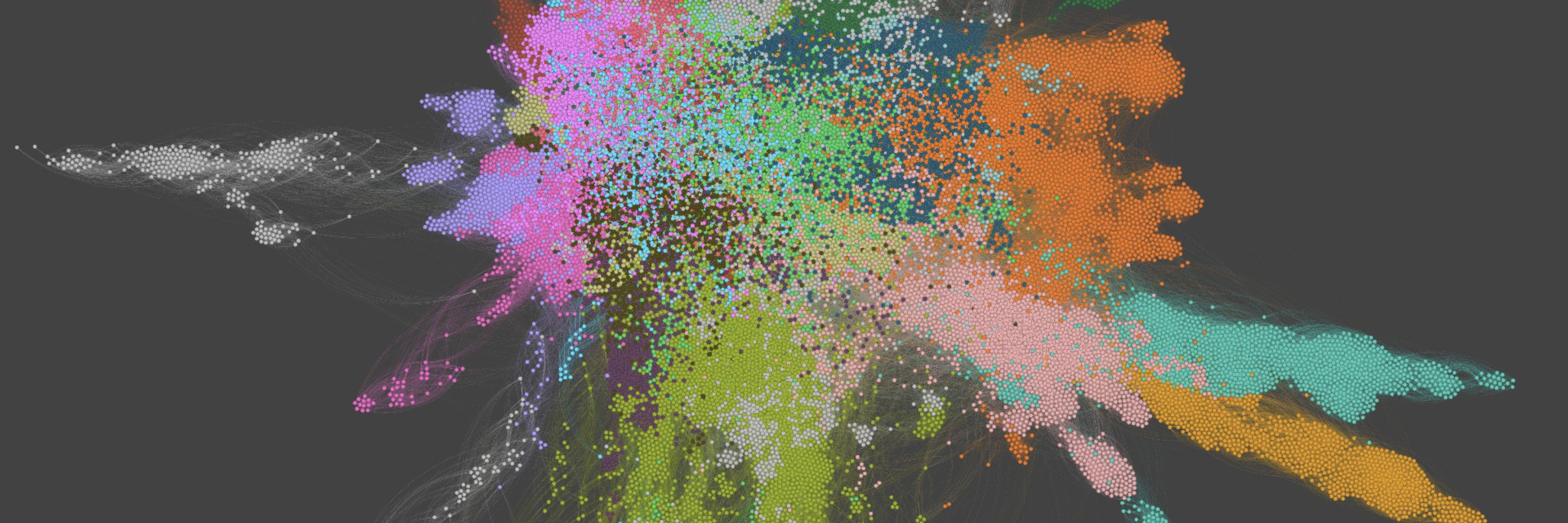
| ML fan & critic | current research mostly #datascience, #machinelearning, #cheminformatics #dataviz #nlp | ✨ #openscience #openaccess #rse | living data point 🚲
GitHub: github.com/matchms/matc...
#opensource #RSE #researchsoftwareengineering

GitHub: github.com/matchms/matc...
#opensource #RSE #researchsoftwareengineering
We also noted the same thing, which triggered us to point out some pitfalls of various fingerprints --> www.biorxiv.org/content/10.1...
We also noted the same thing, which triggered us to point out some pitfalls of various fingerprints --> www.biorxiv.org/content/10.1...
We also highlight options for count fingerprints, such as log-counts and IDF weighted counts. The latter can be used to adjust the bit importance to a dataset of your choice.
An example use-case are chemical space visualizations.
Preprint: www.biorxiv.org/content/10.1...
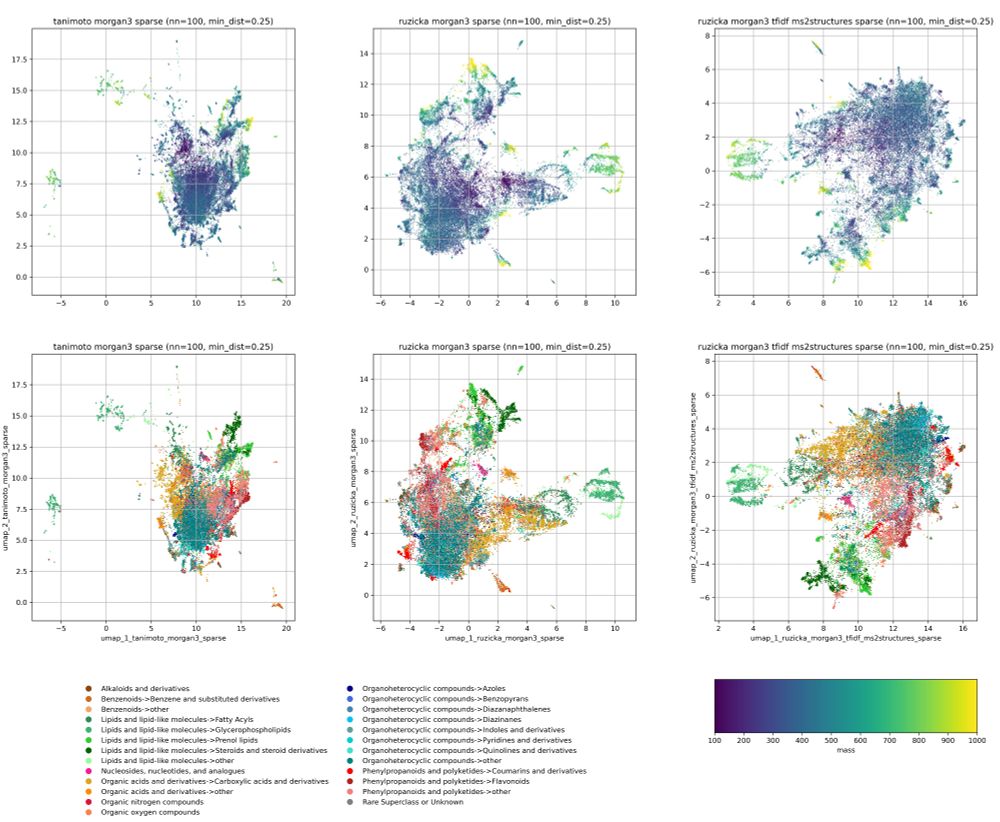
We also highlight options for count fingerprints, such as log-counts and IDF weighted counts. The latter can be used to adjust the bit importance to a dataset of your choice.
An example use-case are chemical space visualizations.
Preprint: www.biorxiv.org/content/10.1...
A huge issue is bit collisions.
Fingerprints with a high bit occupation (RDKit, MAP4) often lead to (1) arbitrary misinterpretations, (2) shifts to high Tanimoto scores, (3) very different handling of small and large molecules.
--> Consider using sparse fingerprints!
--> Morgan >> MAP4 / RDKit
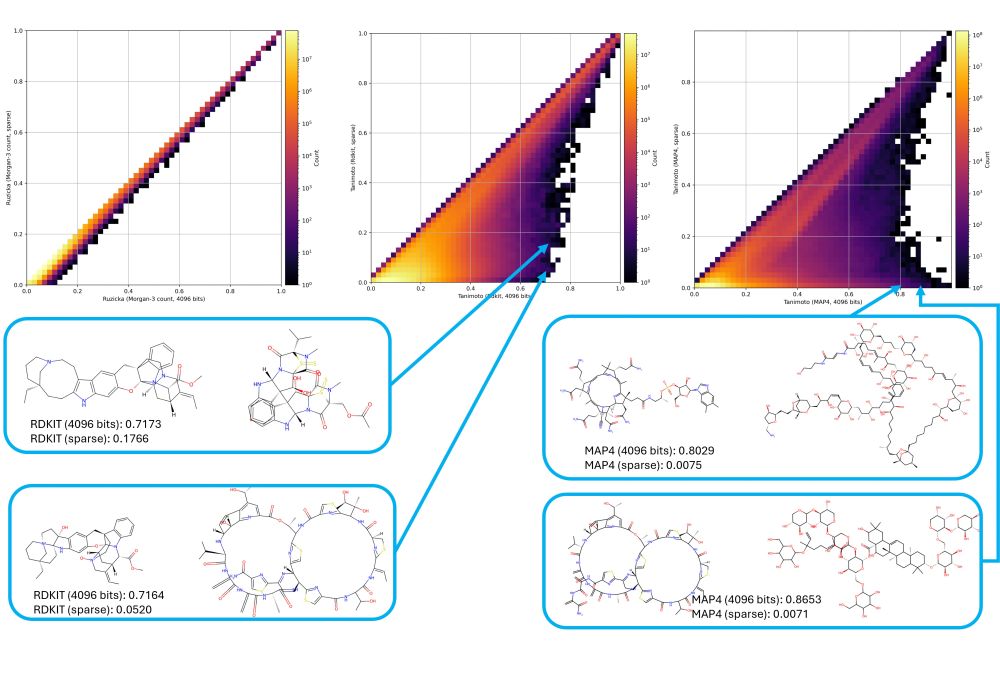
A huge issue is bit collisions.
Fingerprints with a high bit occupation (RDKit, MAP4) often lead to (1) arbitrary misinterpretations, (2) shifts to high Tanimoto scores, (3) very different handling of small and large molecules.
--> Consider using sparse fingerprints!
--> Morgan >> MAP4 / RDKit
We focused on weaknesses of the fingerprints.
Many show frequent duplicates, so same fingerprint for different compounds. Most problematic: this can include *very* different compounds ending up with identical fingerprints.
- MAP4 >> Morgan-type >> daylight
- count >> binary
#cheminformatics
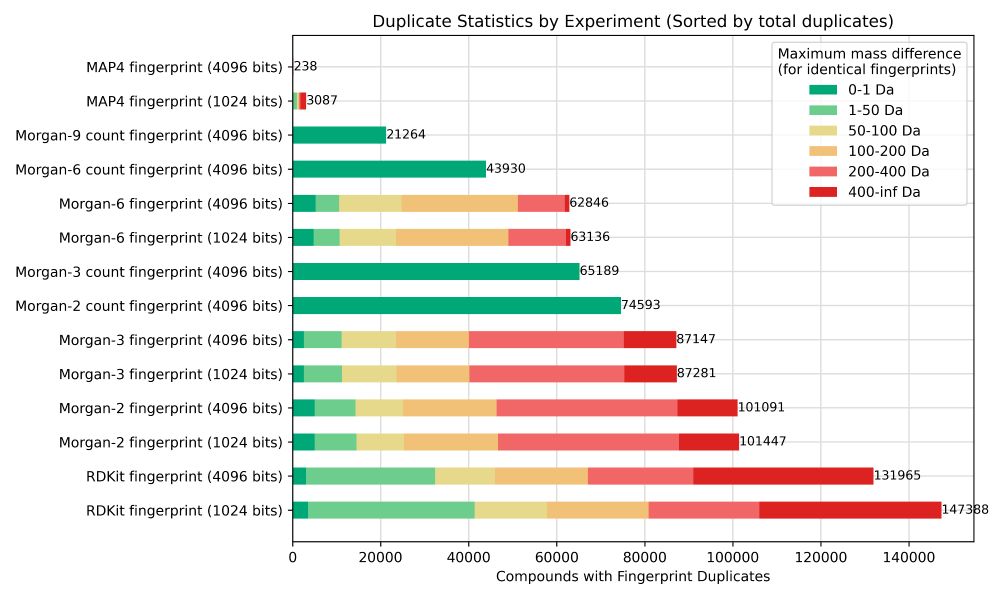
We focused on weaknesses of the fingerprints.
Many show frequent duplicates, so same fingerprint for different compounds. Most problematic: this can include *very* different compounds ending up with identical fingerprints.
- MAP4 >> Morgan-type >> daylight
- count >> binary
#cheminformatics

