| ML fan & critic | current research mostly #datascience, #machinelearning, #cheminformatics #dataviz #nlp | ✨ #openscience #openaccess #rse | living data point 🚲
With functionalities that were on our TODO list for a looooong time: Flash Entropy and BLINK scores! The new "FlashSimilarity" allows computing modified cosine, spectral entropy etc., about 100x faster (or more if you use Linux).
#Python #opensource #massspec

With functionalities that were on our TODO list for a looooong time: Flash Entropy and BLINK scores! The new "FlashSimilarity" allows computing modified cosine, spectral entropy etc., about 100x faster (or more if you use Linux).
#Python #opensource #massspec
#MetSoc25
I am super glad they now also provide options to combine with #Python and #matchms (thanks🙏)



#MetSoc25
I am super glad they now also provide options to combine with #Python and #matchms (thanks🙏)
Glad to also spot #matchms in this universe :)


Glad to also spot #matchms in this universe :)
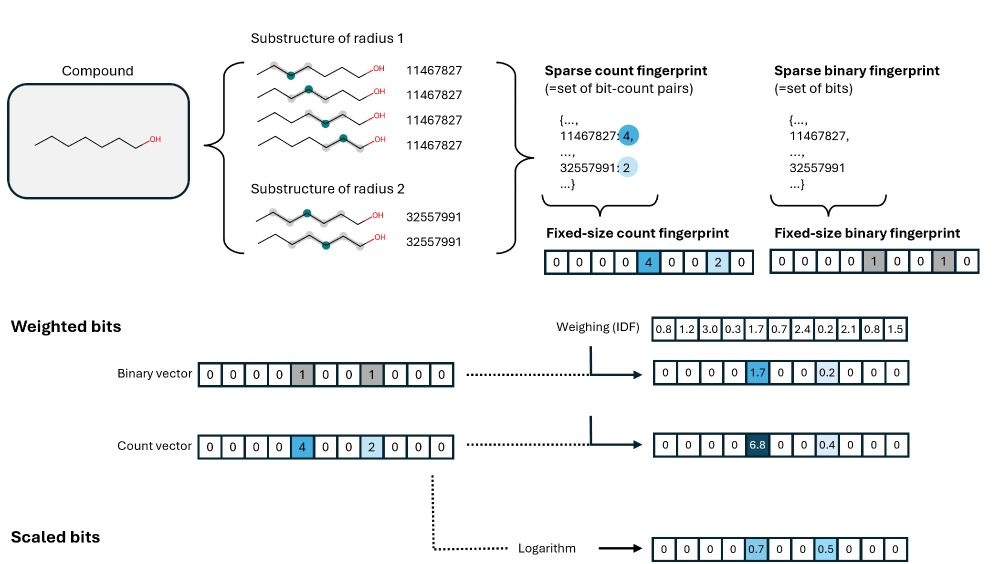
We also highlight options for count fingerprints, such as log-counts and IDF weighted counts. The latter can be used to adjust the bit importance to a dataset of your choice.
An example use-case are chemical space visualizations.
Preprint: www.biorxiv.org/content/10.1...
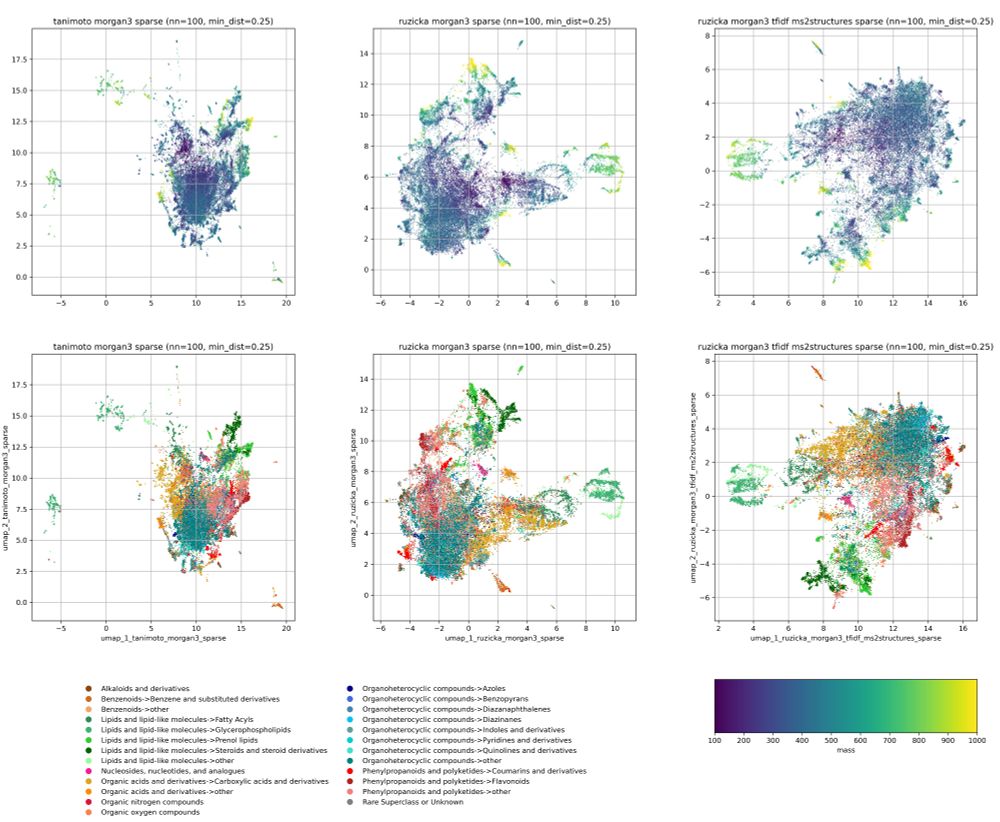
We also highlight options for count fingerprints, such as log-counts and IDF weighted counts. The latter can be used to adjust the bit importance to a dataset of your choice.
An example use-case are chemical space visualizations.
Preprint: www.biorxiv.org/content/10.1...
A huge issue is bit collisions.
Fingerprints with a high bit occupation (RDKit, MAP4) often lead to (1) arbitrary misinterpretations, (2) shifts to high Tanimoto scores, (3) very different handling of small and large molecules.
--> Consider using sparse fingerprints!
--> Morgan >> MAP4 / RDKit
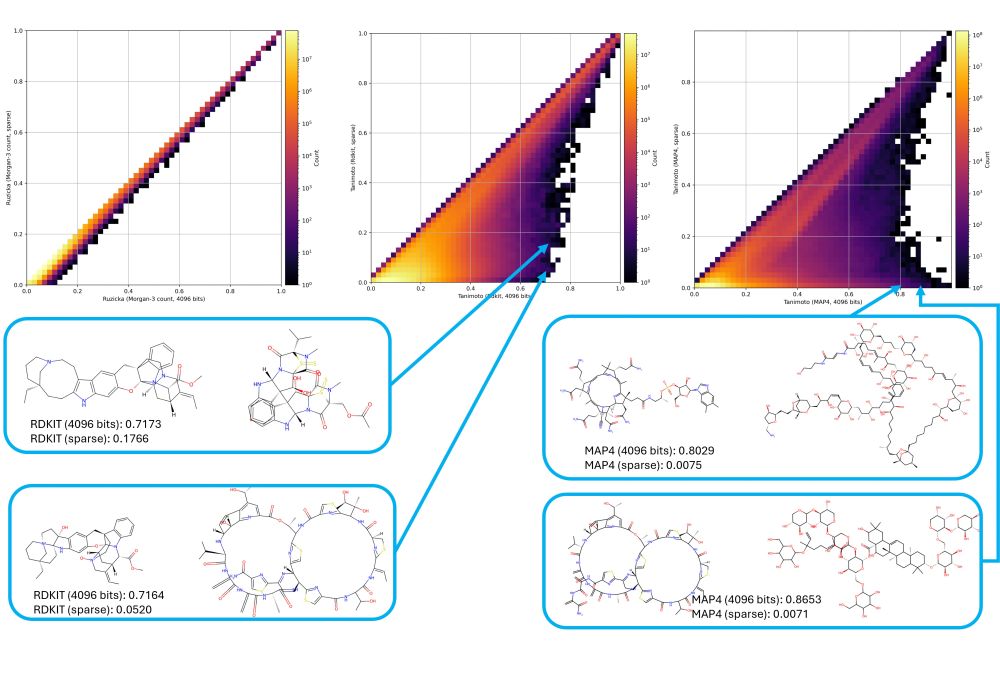
A huge issue is bit collisions.
Fingerprints with a high bit occupation (RDKit, MAP4) often lead to (1) arbitrary misinterpretations, (2) shifts to high Tanimoto scores, (3) very different handling of small and large molecules.
--> Consider using sparse fingerprints!
--> Morgan >> MAP4 / RDKit
We focused on weaknesses of the fingerprints.
Many show frequent duplicates, so same fingerprint for different compounds. Most problematic: this can include *very* different compounds ending up with identical fingerprints.
- MAP4 >> Morgan-type >> daylight
- count >> binary
#cheminformatics
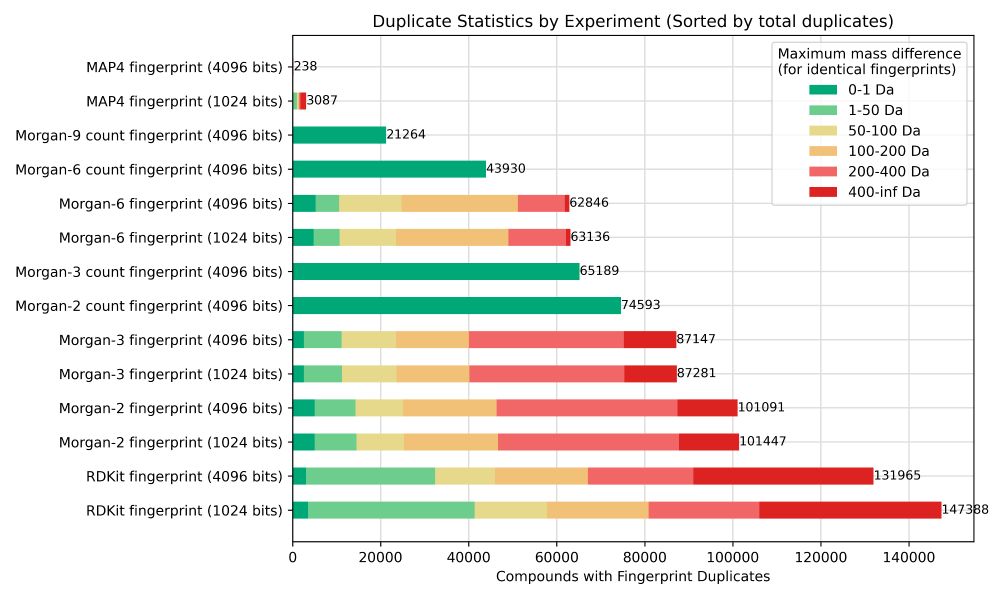
We focused on weaknesses of the fingerprints.
Many show frequent duplicates, so same fingerprint for different compounds. Most problematic: this can include *very* different compounds ending up with identical fingerprints.
- MAP4 >> Morgan-type >> daylight
- count >> binary
#cheminformatics
1/4
@julianpollmann.bsky.social and I went down several rabbit holes to assess some commonly used molecular fingerprints.
Bottom line: For large datasets, make an effort to select suitable settings. "We used Tanimoto" is not good enough.
--> www.biorxiv.org/content/10.1...
1/4
@julianpollmann.bsky.social and I went down several rabbit holes to assess some commonly used molecular fingerprints.
Bottom line: For large datasets, make an effort to select suitable settings. "We used Tanimoto" is not good enough.
--> www.biorxiv.org/content/10.1...

These are the kind of moments that remind me how great the European project is. No border controls, no visas. Just a train following a river to the neighboring country.

These are the kind of moments that remind me how great the European project is. No border controls, no visas. Just a train following a river to the neighboring country.
#Düsseldorf weiterhin konstant bei 4- im #ADFC Klimatest. Läuft. @adfcnrw.bsky.social @adfc-duesseldorf.de
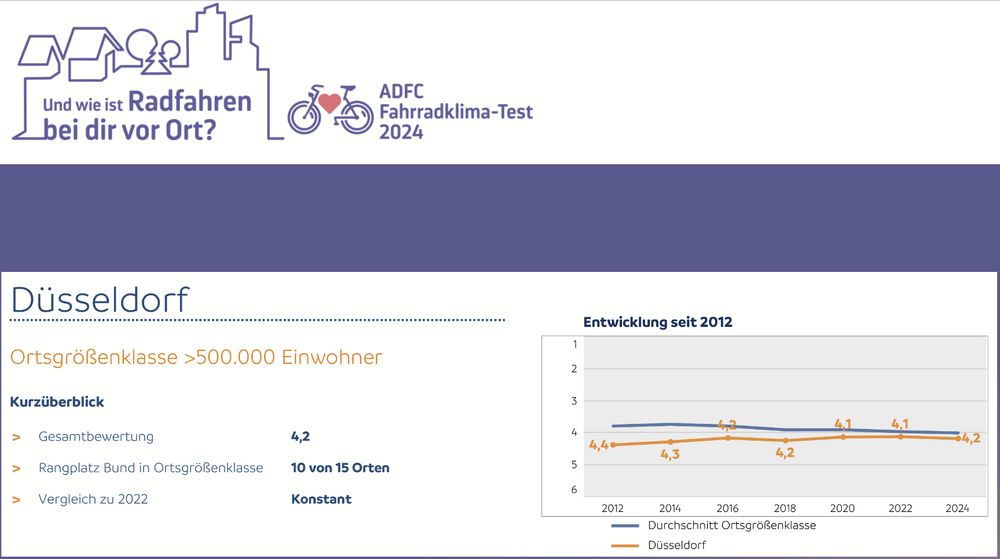
#Düsseldorf weiterhin konstant bei 4- im #ADFC Klimatest. Läuft. @adfcnrw.bsky.social @adfc-duesseldorf.de
(added some text edits and more sketches/figures to the NLP chapters of the "Hands-on Introduction to #DataScience with #Python" textbook)
florian-huber.github.io/data_science...
#OpenScience #Teaching #CCBY
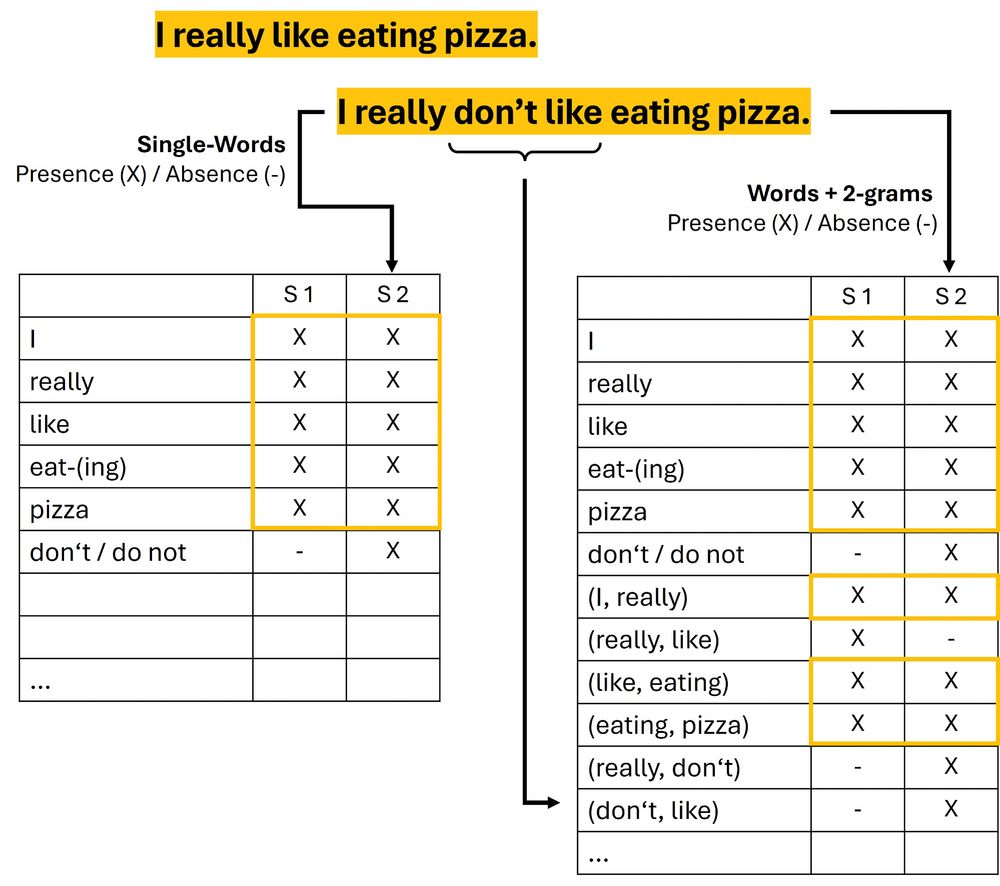
(added some text edits and more sketches/figures to the NLP chapters of the "Hands-on Introduction to #DataScience with #Python" textbook)
florian-huber.github.io/data_science...
#OpenScience #Teaching #CCBY
Contains many text edits and figure updates. For instance, in the sections on Clustering and Machine Learning.
All fully #opensource and #openaccess. Figures are #CCBY.
--> florian-huber.github.io/data_science...
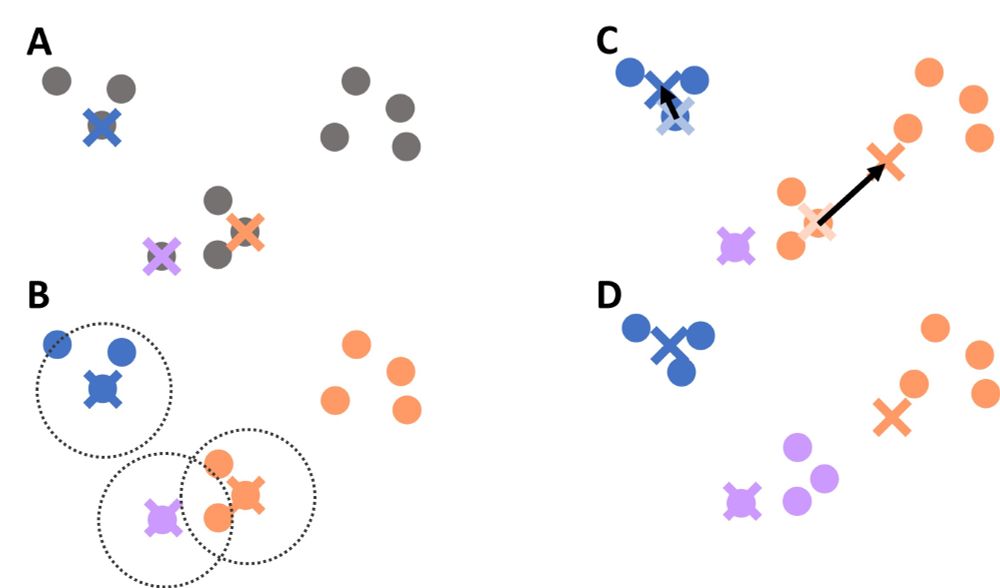
Contains many text edits and figure updates. For instance, in the sections on Clustering and Machine Learning.
All fully #opensource and #openaccess. Figures are #CCBY.
--> florian-huber.github.io/data_science...

--> medium.com/@f.huber/wat...
#DataScience #Python #Teaching

--> medium.com/@f.huber/wat...
#DataScience #Python #Teaching
#opensource #Python #massspec

#opensource #Python #massspec
#microtubules with EB3 growing against rigid barriers.
--> www.cell.com/biophysj/ful...

#microtubules with EB3 growing against rigid barriers.
--> www.cell.com/biophysj/ful...
--> aurora.ndimensional.xyz

--> aurora.ndimensional.xyz
--> royalsocietypublishing.org/doi/10.1098/...
#OpenAccess #DataScience

--> royalsocietypublishing.org/doi/10.1098/...
#OpenAccess #DataScience
--> florian-huber.github.io/python-intro...
#DataScience #teaching

--> florian-huber.github.io/python-intro...
#DataScience #teaching
The content is now mostly complete. Text and figures will undergo further polishing.
--> florian-huber.github.io/data_science...
#datascience #opensource #teaching

The content is now mostly complete. Text and figures will undergo further polishing.
--> florian-huber.github.io/data_science...
#datascience #opensource #teaching


