

www.cell.com/cell-reports...
www.cell.com/cell-reports...
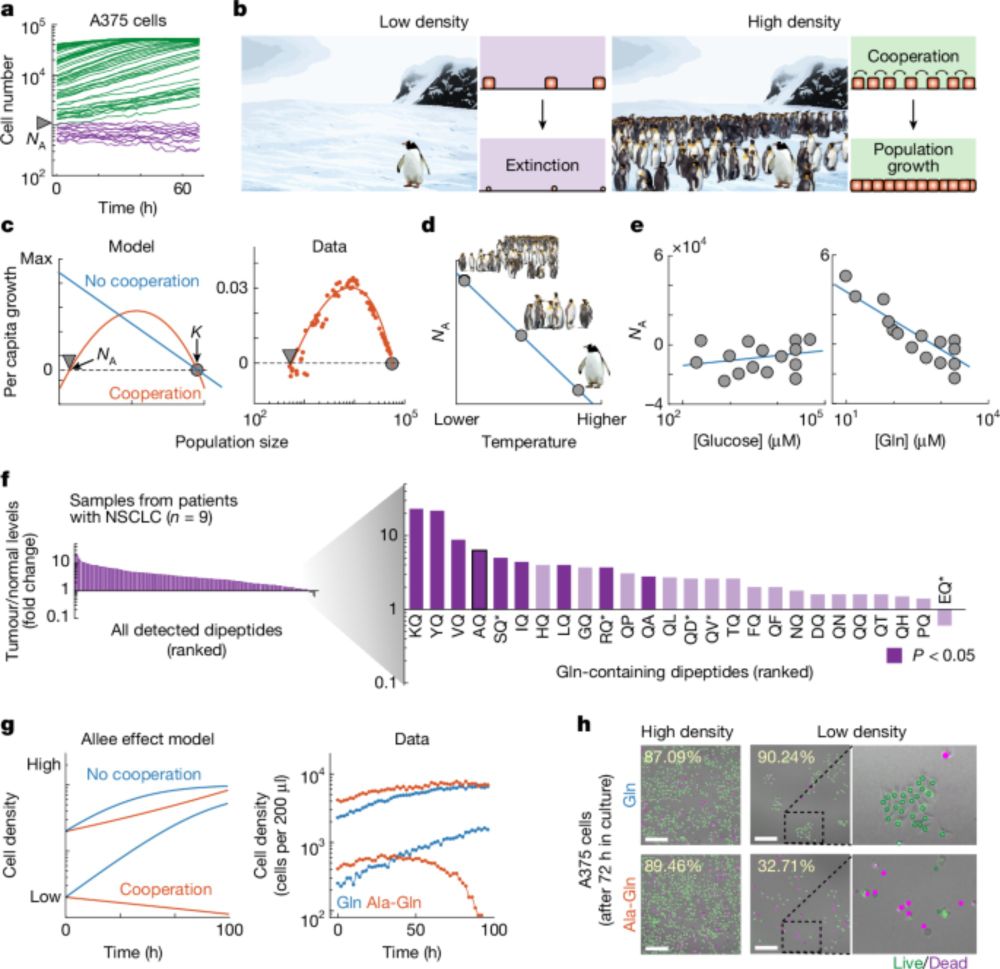
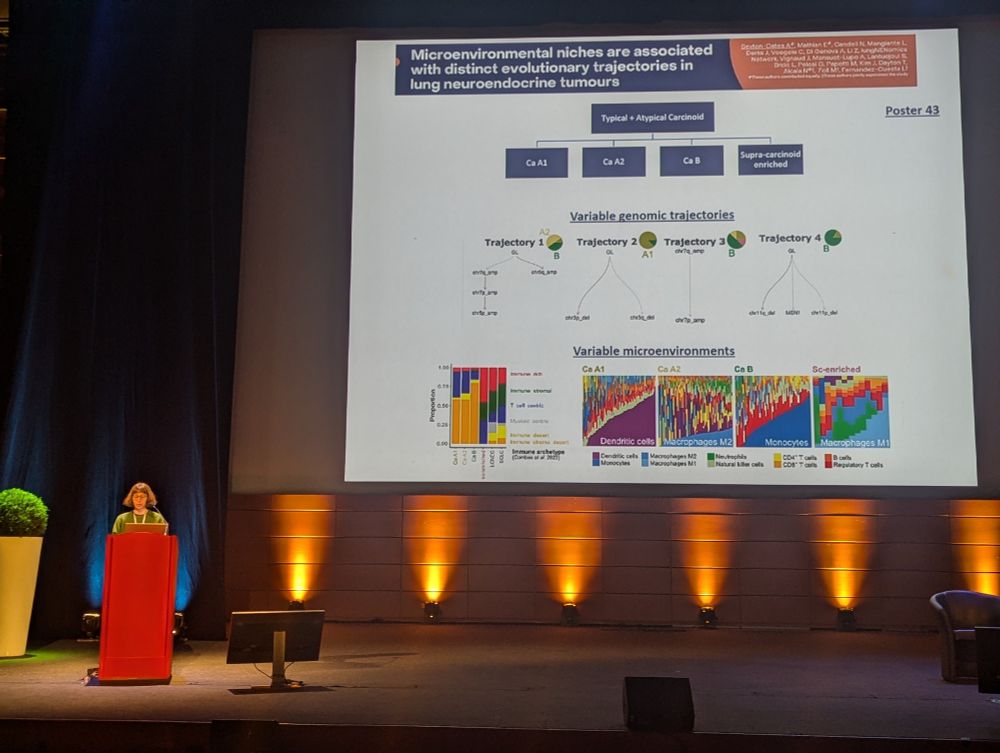
We are very pleased to introduce #StrainDifferences: “Genetic background sets the trajectory of cancer evolution”
www.biorxiv.org/content/10.1...
🧵[1/12]

We are very pleased to introduce #StrainDifferences: “Genetic background sets the trajectory of cancer evolution”
www.biorxiv.org/content/10.1...
🧵[1/12]
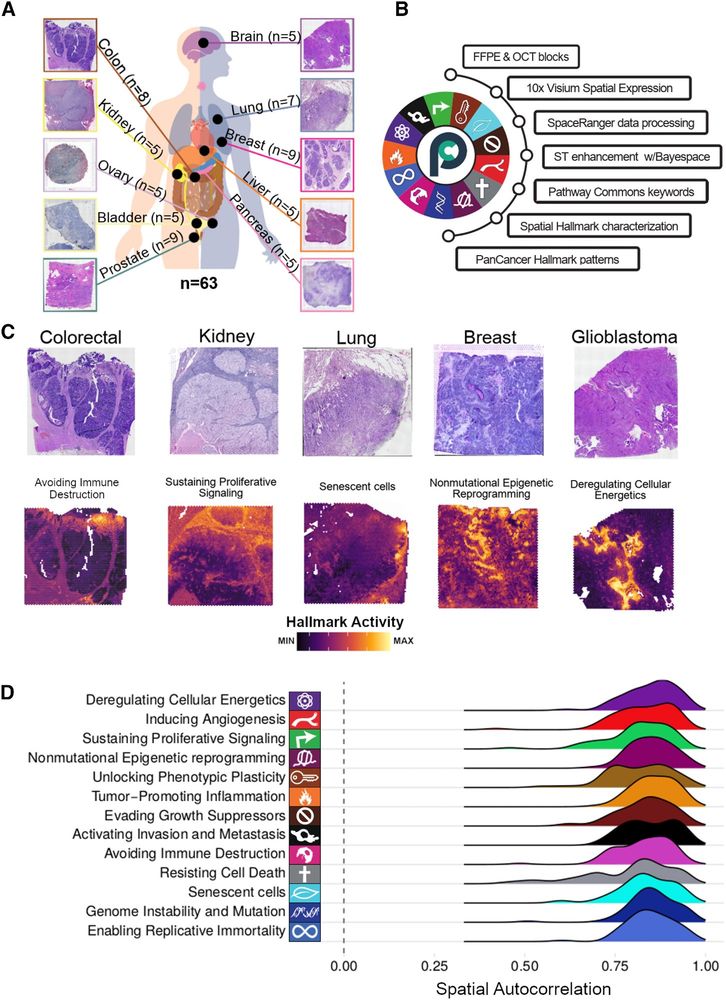
Original work 👉 doi.org/10.1038/s415...
Our Commentary 👇
Original work 👉 doi.org/10.1038/s415...
Our Commentary 👇
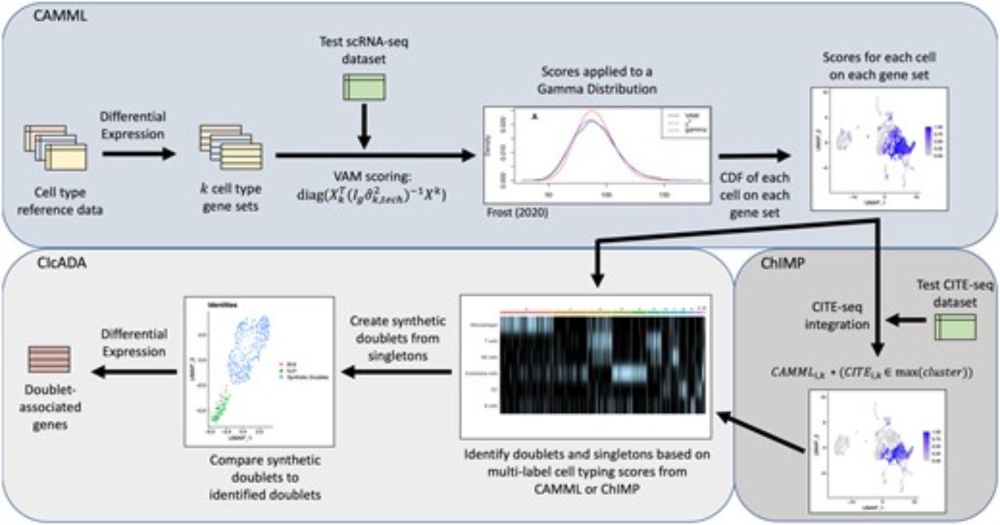
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
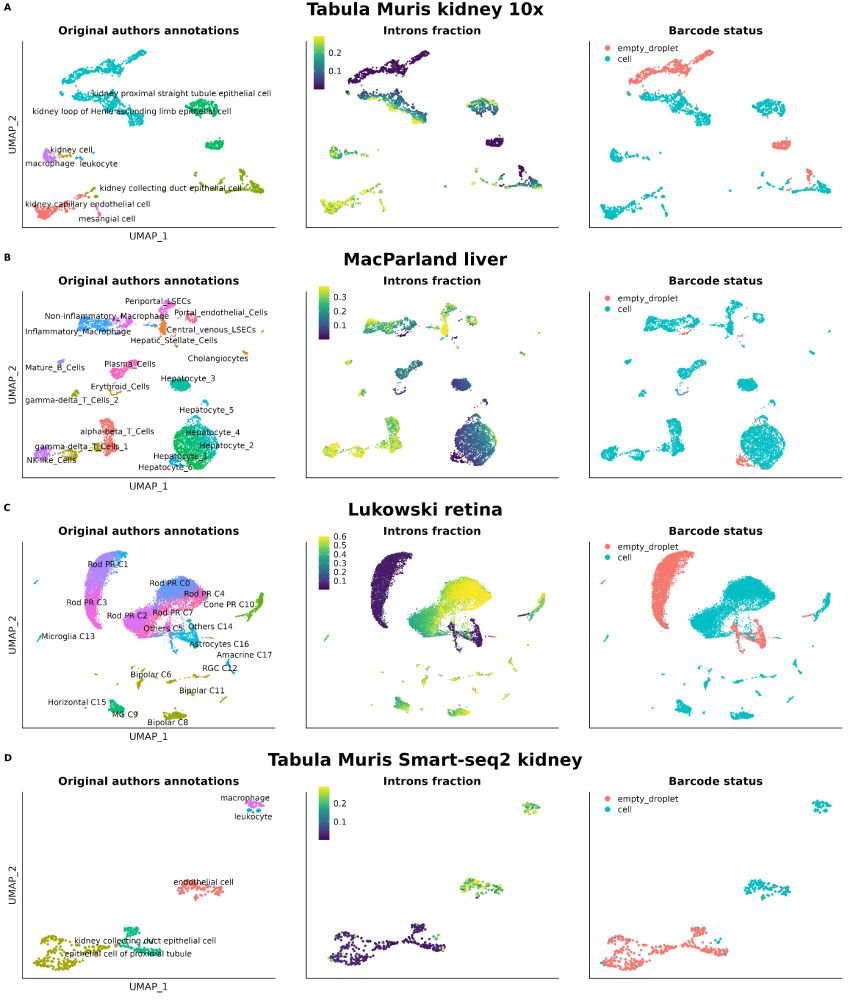
Not checking nuclear markers like MALAT1 or intronic reads in your scRNA-seq data?🚨
We show their power to flag low-quality cells—even in top public datasets. It’s time to prioritize better QC for cleaner, more reliable genomics research!
Read more: bmcgenomics.biomedcentral.com/articles/10....
1/8
#PostdocJobs https://x.com/CancersRare/status/1726698337007776217
#PostdocJobs https://x.com/CancersRare/status/1726698337007776217
#PostdocJobs https://x.com/CancersRare/status/1726697028372348929
#PostdocJobs https://x.com/CancersRare/status/1726697028372348929