
Matthew Feickert
@matthewfeickert.com
Research scientist at UW-Madison data science institute working on #LHC #physics and data science with the ATLAS experiment @ CERN and IRIS-HEP. PhD @ SMU
Seems that #pyhf has finally creeped over the 200 citations on INSPIRE recently. :) 🎉
inspirehep.net/literature?q...
inspirehep.net/literature/1...
Nice little milestone for us all to celebrate, @lukasheinrich.com, @giordonstark.com, @kylecranmer.bsky.social. Cheers!
inspirehep.net/literature?q...
inspirehep.net/literature/1...
Nice little milestone for us all to celebrate, @lukasheinrich.com, @giordonstark.com, @kylecranmer.bsky.social. Cheers!

October 9, 2025 at 12:39 PM
Seems that #pyhf has finally creeped over the 200 citations on INSPIRE recently. :) 🎉
inspirehep.net/literature?q...
inspirehep.net/literature/1...
Nice little milestone for us all to celebrate, @lukasheinrich.com, @giordonstark.com, @kylecranmer.bsky.social. Cheers!
inspirehep.net/literature?q...
inspirehep.net/literature/1...
Nice little milestone for us all to celebrate, @lukasheinrich.com, @giordonstark.com, @kylecranmer.bsky.social. Cheers!
I noticed today that PyPI now (not sure for how long) has a filtering option when you look at the distributions of packages. That's a nice quality of life improvement.

September 26, 2025 at 9:56 PM
I noticed today that PyPI now (not sure for how long) has a filtering option when you look at the distributions of packages. That's a nice quality of life improvement.
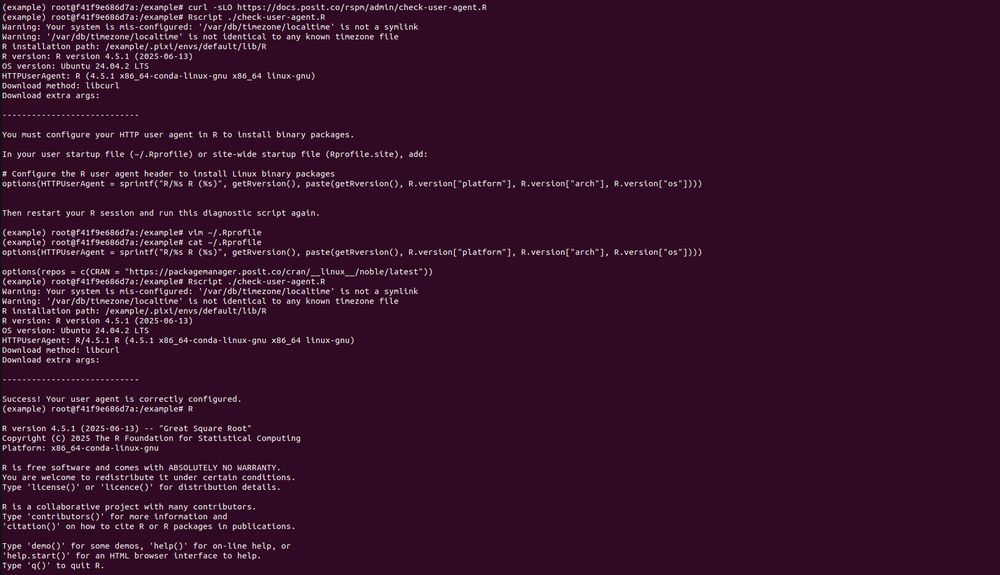
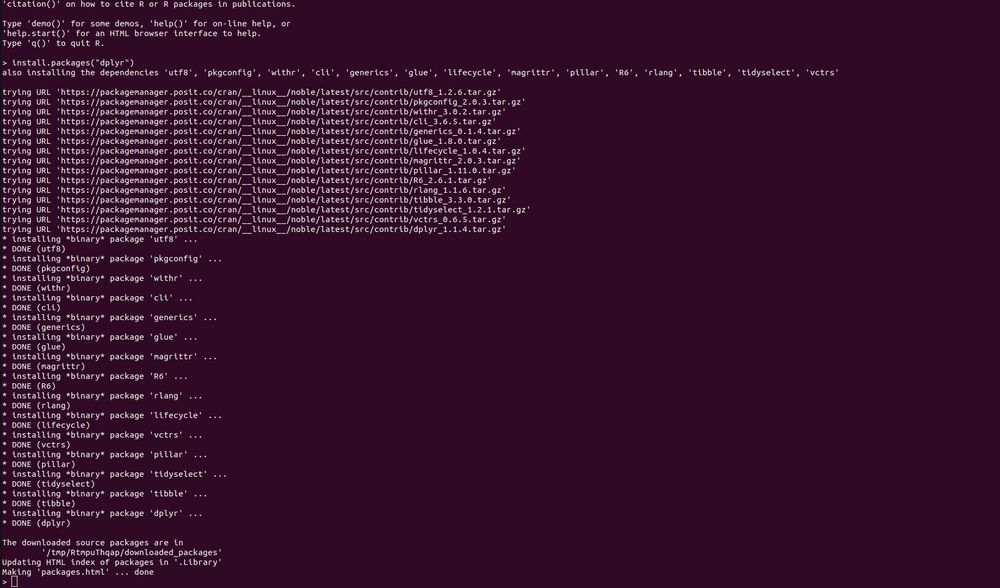
Thanks for the detailed summary, as that combination of things works. So if I understand things correctly, this is the required way to do things _now_, but whenever the next release of Package Manager happens there will be new docs on how to just provide an index URL in ~/.Rprofile to get binaries?
July 23, 2025 at 5:19 AM
Thanks for the detailed summary, as that combination of things works. So if I understand things correctly, this is the required way to do things _now_, but whenever the next release of Package Manager happens there will be new docs on how to just provide an index URL in ~/.Rprofile to get binaries?
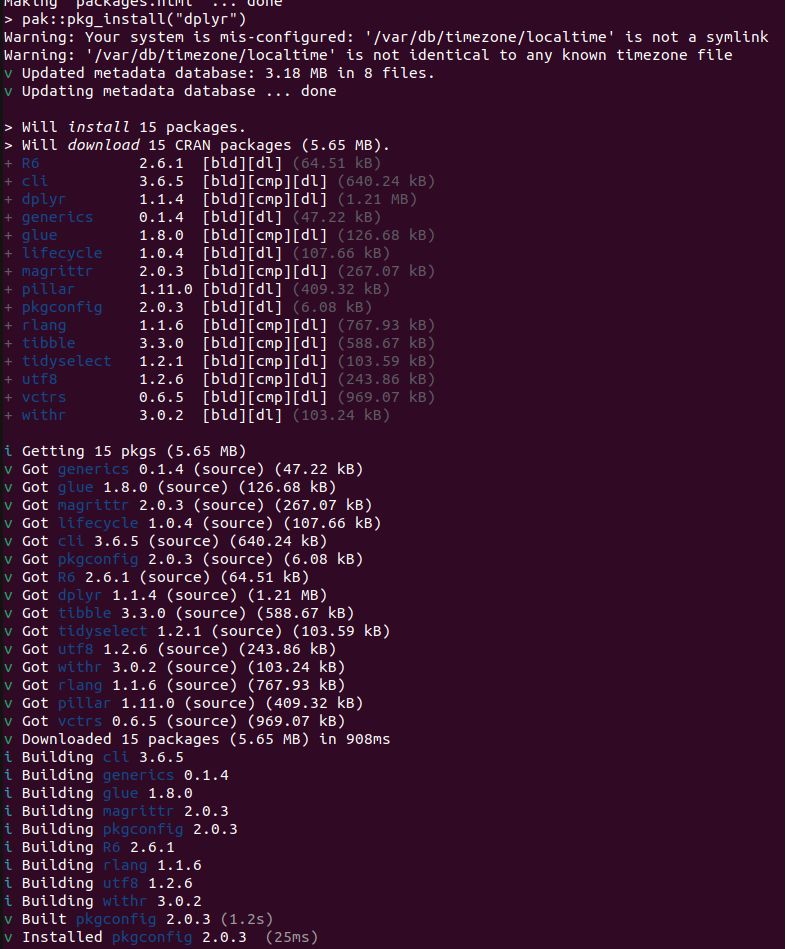
* What does
```
pak::pkg_install("dplyr")
```
do? It builds 15 packages from source. This is for a Ubuntu 22.04 x86 Linux container with R v4.5.x. So pak is not getting built distributions, because they don't exist. This is not a solution.
```
pak::pkg_install("dplyr")
```
do? It builds 15 packages from source. This is for a Ubuntu 22.04 x86 Linux container with R v4.5.x. So pak is not getting built distributions, because they don't exist. This is not a solution.
July 23, 2025 at 3:02 AM
* What does
```
pak::pkg_install("dplyr")
```
do? It builds 15 packages from source. This is for a Ubuntu 22.04 x86 Linux container with R v4.5.x. So pak is not getting built distributions, because they don't exist. This is not a solution.
```
pak::pkg_install("dplyr")
```
do? It builds 15 packages from source. This is for a Ubuntu 22.04 x86 Linux container with R v4.5.x. So pak is not getting built distributions, because they don't exist. This is not a solution.
Example: To add bioconda.github.io to the channel logic (with a priority lower than conda-forge)
```
pixi workspace channel add bioconda
```
now means that your Pixi manifest "knows" about every package on Bioconda and can solve against it _and_ conda-forge.
```
pixi workspace channel add bioconda
```
now means that your Pixi manifest "knows" about every package on Bioconda and can solve against it _and_ conda-forge.

July 23, 2025 at 2:00 AM
Example: To add bioconda.github.io to the channel logic (with a priority lower than conda-forge)
```
pixi workspace channel add bioconda
```
now means that your Pixi manifest "knows" about every package on Bioconda and can solve against it _and_ conda-forge.
```
pixi workspace channel add bioconda
```
now means that your Pixi manifest "knows" about every package on Bioconda and can solve against it _and_ conda-forge.
To answer your question about "where the binary packages are coming from", I'll give the short answer and then a longer one.
Short answer: Run `pixi list`
This will show multiple columns of information including "source", which shows things are coming from conda-forge. conda-forge.org/packages/
Short answer: Run `pixi list`
This will show multiple columns of information including "source", which shows things are coming from conda-forge. conda-forge.org/packages/

July 23, 2025 at 2:00 AM
To answer your question about "where the binary packages are coming from", I'll give the short answer and then a longer one.
Short answer: Run `pixi list`
This will show multiple columns of information including "source", which shows things are coming from conda-forge. conda-forge.org/packages/
Short answer: Run `pixi list`
This will show multiple columns of information including "source", which shows things are coming from conda-forge. conda-forge.org/packages/
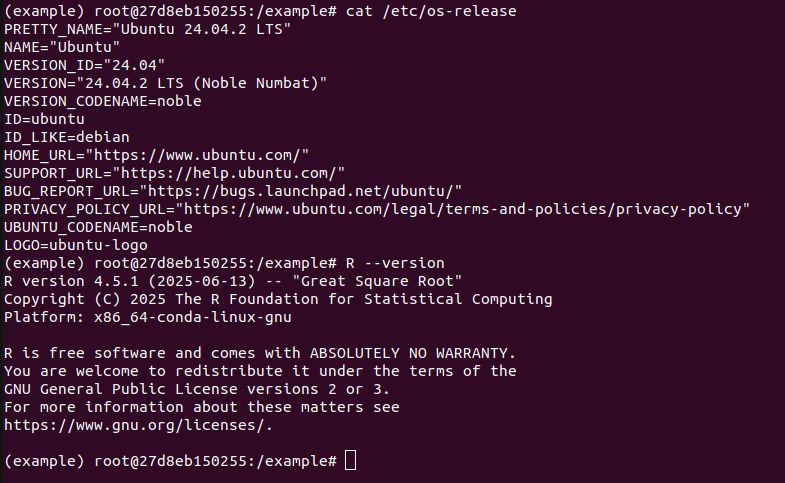
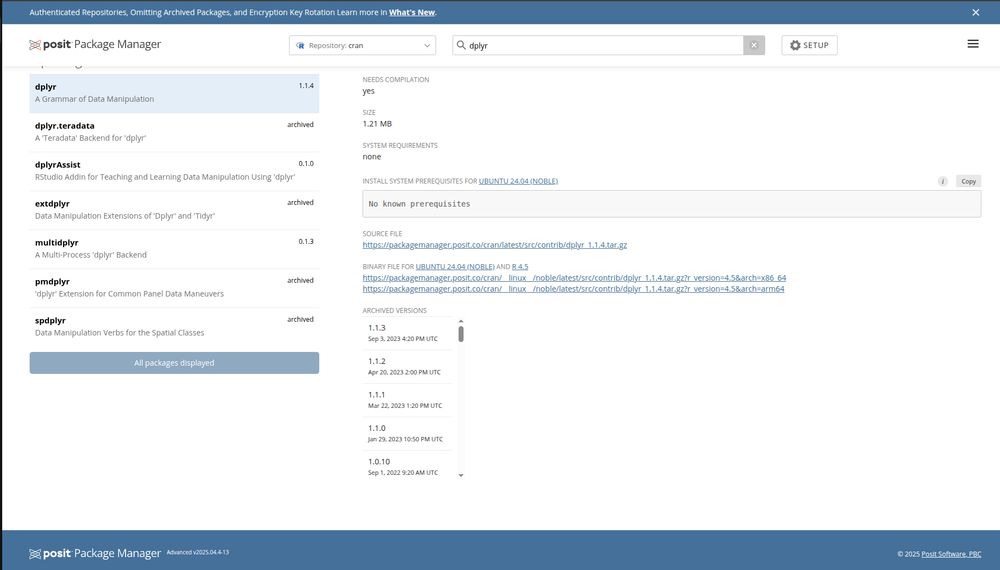
Is a Ubuntu 22.04 Linux container and R v4.5.x far from the usual ecosystem? I don't see how that can realistically be the case. Following the docs it goes right for the source distribution.
* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...


July 22, 2025 at 10:07 PM
Is a Ubuntu 22.04 Linux container and R v4.5.x far from the usual ecosystem? I don't see how that can realistically be the case. Following the docs it goes right for the source distribution.
* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...
Hi Hadley. Thanks for taking time to comment. TIL that Posit Public Package Manager exists, I had only known about posit package manager. Though regardless, what is the benefit if it doesn't actually provide binaries? packagemanager.posit.co/client/#/rep...

July 22, 2025 at 3:38 PM
Hi Hadley. Thanks for taking time to comment. TIL that Posit Public Package Manager exists, I had only known about posit package manager. Though regardless, what is the benefit if it doesn't actually provide binaries? packagemanager.posit.co/client/#/rep...
What is your workflow with Posit package manager like? I am not an expert user, but the UX seems absurdly bad as presented. Maybe this is what R users are used to expecting, but if from the command line I can't install things that seems very broken. Also, it just doesn't work?


July 22, 2025 at 3:23 PM
What is your workflow with Posit package manager like? I am not an expert user, but the UX seems absurdly bad as presented. Maybe this is what R users are used to expecting, but if from the command line I can't install things that seems very broken. Also, it just doesn't work?
Happy #HiggspendenceDay to everyone! It has truly been an amazing period of knowledge growth since the discovery announcement. :)

July 4, 2025 at 10:38 PM
Happy #HiggspendenceDay to everyone! It has truly been an amazing period of knowledge growth since the discovery announcement. :)
After listening to her amazing music for well over a decade, I had the wonderful opportunity to see @zoekeating.com perform live in Boulder, CO tonight! Such a joy and very moving and powerful evening. Also, learned about some excellent documentaries to check out that she’s composed for!


May 16, 2025 at 5:23 AM
After listening to her amazing music for well over a decade, I had the wonderful opportunity to see @zoekeating.com perform live in Boulder, CO tonight! Such a joy and very moving and powerful evening. Also, learned about some excellent documentaries to check out that she’s composed for!
From my non-Bluesky friend on our Discord

May 9, 2025 at 3:56 AM
From my non-Bluesky friend on our Discord
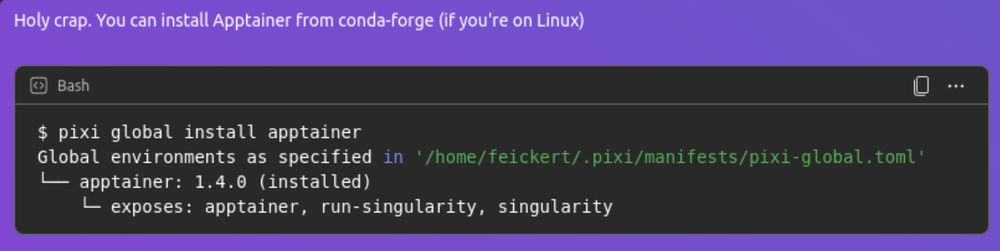
While I'm grump about it, I have to give props to Microsoft Teams for having the nicest code box with language formatting experience of any chat client that I've used.
May 6, 2025 at 11:08 PM
While I'm grump about it, I have to give props to Microsoft Teams for having the nicest code box with language formatting experience of any chat client that I've used.
I’m joining about 80 of my colleagues this week to meet with congressional offices, as private citizens and scientists, to advocate for continued investment in basic scientific research and supporting particle physics research. There’s urgency to the message. #HEPOnTheHill2025

April 22, 2025 at 3:59 PM
I’m joining about 80 of my colleagues this week to meet with congressional offices, as private citizens and scientists, to advocate for continued investment in basic scientific research and supporting particle physics research. There’s urgency to the message. #HEPOnTheHill2025
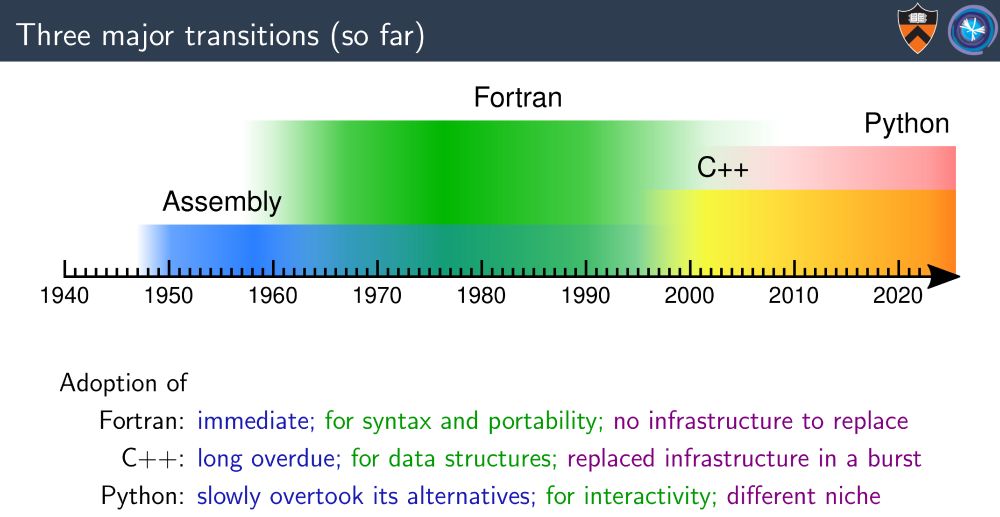
Particle physics is mostly still custom C++ with then end user analysis done in Python. Though as a field this is somewhat recent, as we only moved from Fortran to C++ in the early 2000s.
(Credit to my colleague Jim Pivarski whose talk I took the image from: indico.jlab.org/event/505/co...)
(Credit to my colleague Jim Pivarski whose talk I took the image from: indico.jlab.org/event/505/co...)
April 17, 2025 at 7:20 PM
Particle physics is mostly still custom C++ with then end user analysis done in Python. Though as a field this is somewhat recent, as we only moved from Fortran to C++ in the early 2000s.
(Credit to my colleague Jim Pivarski whose talk I took the image from: indico.jlab.org/event/505/co...)
(Credit to my colleague Jim Pivarski whose talk I took the image from: indico.jlab.org/event/505/co...)
#TIL from my DSI colleagues about the undocumented `seaborn.dogplot`. Amazing!
github.com/mwaskom/seab...
github.com/mwaskom/seab...

April 10, 2025 at 6:48 PM
#TIL from my DSI colleagues about the undocumented `seaborn.dogplot`. Amazing!
github.com/mwaskom/seab...
github.com/mwaskom/seab...
Still trying to learn to pour designs symmetrically (also got a few too many bigger bubbles in the microfoam). Tastes amazing though!

March 25, 2025 at 3:23 AM
Still trying to learn to pour designs symmetrically (also got a few too many bigger bubbles in the microfoam). Tastes amazing though!
Also always great to get to work with @lukasheinrich.com, one of the people who set a new vision for our fields’ computing tools, discussing Snakemake + REANA plans with @tiborsimko.bsky.social and thinking about what future analysis techniques and technologies our work this week can bring!

March 15, 2025 at 10:34 AM
Also always great to get to work with @lukasheinrich.com, one of the people who set a new vision for our fields’ computing tools, discussing Snakemake + REANA plans with @tiborsimko.bsky.social and thinking about what future analysis techniques and technologies our work this week can bring!
I really enjoyed getting to collaborate with my CMS and LHCb colleagues, @clelange.bsky.social and Chris. Looking forward to keeping this momentum going with some experiments with software deployment in the near future.

March 15, 2025 at 9:21 AM
I really enjoyed getting to collaborate with my CMS and LHCb colleagues, @clelange.bsky.social and Chris. Looking forward to keeping this momentum going with some experiments with software deployment in the near future.
A release made with a live audience. :)

March 14, 2025 at 11:42 PM
A release made with a live audience. :)
I’m not in the photo as I was the one taking it. :P

March 11, 2025 at 12:27 AM
I’m not in the photo as I was the one taking it. :P
Today was a good day to take vacation time and join the #StandUpForScience Denver demonstration. Federally funded science has improved every aspect of human life at an astonishingly good ROI. Defunding science is an attack on knowledge, health, and human life. Being silent is not an option.




March 7, 2025 at 11:35 PM
Today was a good day to take vacation time and join the #StandUpForScience Denver demonstration. Federally funded science has improved every aspect of human life at an astonishingly good ROI. Defunding science is an attack on knowledge, health, and human life. Being silent is not an option.
I gave a talk today in the LHC Reinterpretation Forum urging my particle physics colleagues to get on board with using @conda-forge.org and @prefix.dev's pixi, so very fortuitous that as more of them will be trying things out they're have an even better experience! indico.cern.ch/event/146610...

February 28, 2025 at 2:59 AM
I gave a talk today in the LHC Reinterpretation Forum urging my particle physics colleagues to get on board with using @conda-forge.org and @prefix.dev's pixi, so very fortuitous that as more of them will be trying things out they're have an even better experience! indico.cern.ch/event/146610...
Writing, and revising, fuel.

February 20, 2025 at 4:47 AM
Writing, and revising, fuel.
CDS's discussions feature is something that is generally only enabled on internal documents. Here's a screenshot of what the UI looks like for some proceedings I'm preparing for the ATLAS collaboration now. As this is all internal still and I didn't ask for colleagues consent I've redacted parts.

February 6, 2025 at 7:33 AM
CDS's discussions feature is something that is generally only enabled on internal documents. Here's a screenshot of what the UI looks like for some proceedings I'm preparing for the ATLAS collaboration now. As this is all internal still and I didn't ask for colleagues consent I've redacted parts.

