Our ultra deep sequencing is now published in GigaScience: academic.oup.com/gigascience/...
Soil is wild! Thanks for leading the study @canerbagci.bsky.social
#secmet #soil #metagenomics #bacterialdiversity

Our ultra deep sequencing is now published in GigaScience: academic.oup.com/gigascience/...
Soil is wild! Thanks for leading the study @canerbagci.bsky.social
#secmet #soil #metagenomics #bacterialdiversity
#secmet #lassopeptides #syntheticbiology

#secmet #lassopeptides #syntheticbiology
💡 Share your best name idea and win 500€
👉 More info and submission here: docs.google.com/forms/d/1ym9...
#secmet #NaturalProducts #Antibiotics #NamingChallenge #Glycopeptides @gdwantibiotics.bsky.social @marghesosio.bsky.social

💡 Share your best name idea and win 500€
👉 More info and submission here: docs.google.com/forms/d/1ym9...
#secmet #NaturalProducts #Antibiotics #NamingChallenge #Glycopeptides @gdwantibiotics.bsky.social @marghesosio.bsky.social
m.youtube.com/@ZiemertLab
We’ve uploaded short tutorial videos on how to use our tools for genome mining and natural product discovery.
Thanks Semih, @martinaadamek.bsky.social @turgutmesut.bsky.social ! #GenomeMining #SecMet #naturalproducts

m.youtube.com/@ZiemertLab
We’ve uploaded short tutorial videos on how to use our tools for genome mining and natural product discovery.
Thanks Semih, @martinaadamek.bsky.social @turgutmesut.bsky.social ! #GenomeMining #SecMet #naturalproducts
pubs.acs.org/doi/abs/10.1...
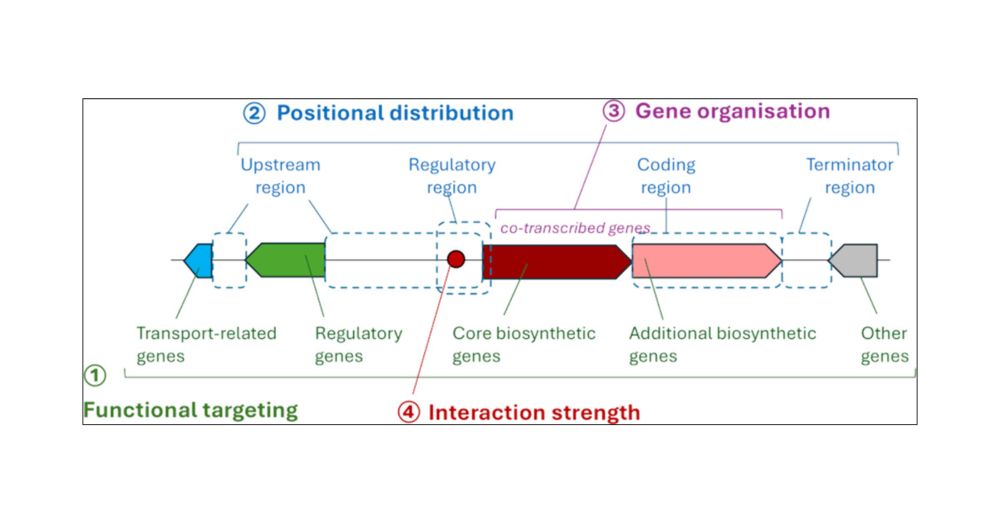
pubs.acs.org/doi/abs/10.1...
Help redefine the #glycopeptide family!
🏆Submit your idea & win 500 € + authorship credit
Let’s rethink antibiotic classification—together!
🧬 Deadline: Sept 15, 2025
👉 docs.google.com/forms/d/e/1F...
@nadineziemert.bsky.social #NameChallenge #NaturalProducts #Antibiotics

Help redefine the #glycopeptide family!
🏆Submit your idea & win 500 € + authorship credit
Let’s rethink antibiotic classification—together!
🧬 Deadline: Sept 15, 2025
👉 docs.google.com/forms/d/e/1F...
@nadineziemert.bsky.social #NameChallenge #NaturalProducts #Antibiotics
pubs.acs.org/doi/10.1021/...


Remarkable work led by @katienoble241.bsky.social and Rebecca Devine demonstrates why antibiotic biosynthesis is often switched on in solid cultures and off in liquid.
www.biorxiv.org/content/10.1...

Remarkable work led by @katienoble241.bsky.social and Rebecca Devine demonstrates why antibiotic biosynthesis is often switched on in solid cultures and off in liquid.
www.biorxiv.org/content/10.1...


