🏳️🌈 he/him


🧵 (11/14)

🧵 (11/14)
🧵 (10/14)

🧵 (10/14)
🧵 (9/14)

🧵 (9/14)
🧵 (8/14)

🧵 (8/14)
🧵 (7/14)

🧵 (7/14)
🧵 (6/14)

🧵 (6/14)
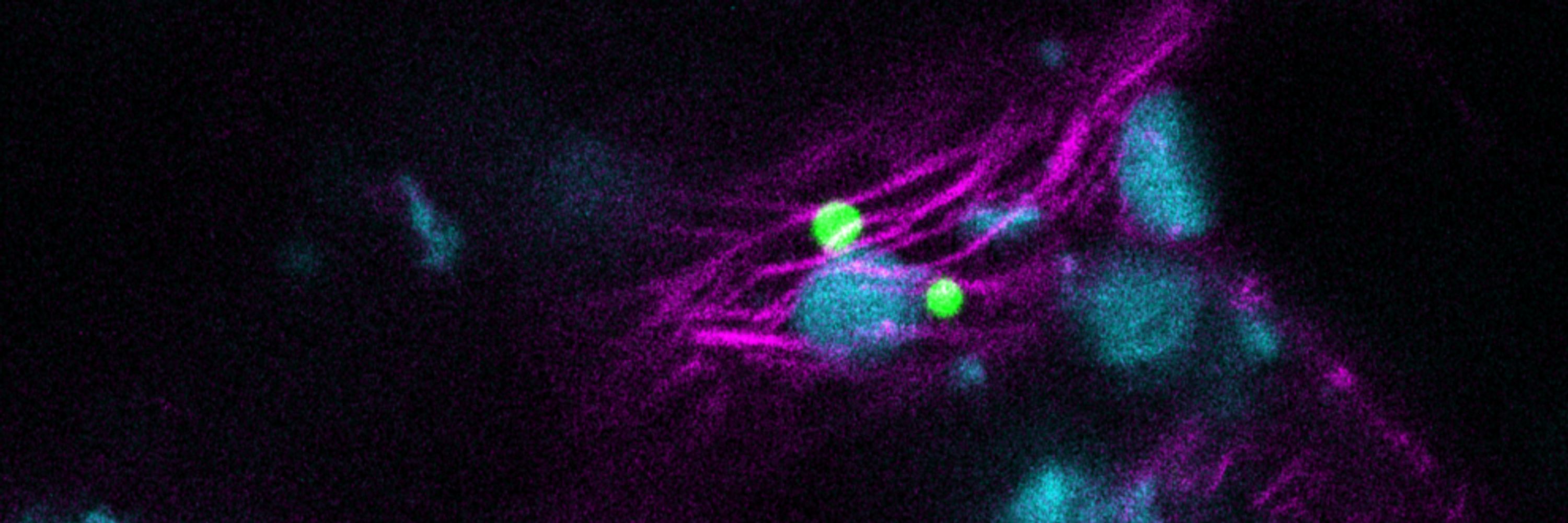
We prove it is rather the second, as the P-body deffective mutant dcp5-1 is more tolerant to Pseudomonas without being majorly affected in canonical defense responses.
🧵 (5/14)

We prove it is rather the second, as the P-body deffective mutant dcp5-1 is more tolerant to Pseudomonas without being majorly affected in canonical defense responses.
🧵 (5/14)
🧵 (4/14)

🧵 (4/14)
🧵 (3/14)

🧵 (3/14)
🧵 (2/14)

🧵 (2/14)
www.biorxiv.org/content/10.1...
🧵 (1/14)

www.biorxiv.org/content/10.1...
🧵 (1/14)

