Machine Learning in Science
@mackelab.bsky.social
We build probabilistic #MachineLearning and #AI Tools for scientific discovery, especially in Neuroscience. Probably not posted by @jakhmack.bsky.social.
📍 @ml4science.bsky.social, Tübingen, Germany
📍 @ml4science.bsky.social, Tübingen, Germany
Congrats to Dr Michael Deistler @deismic.bsky.social, who defended his PhD!
Michael worked on "Machine Learning for Inference in Biophysical Neuroscience Simulations", focusing on simulation-based inference and differentiable simulation.
We wish him all the best for the next chapter! 👏🎓
Michael worked on "Machine Learning for Inference in Biophysical Neuroscience Simulations", focusing on simulation-based inference and differentiable simulation.
We wish him all the best for the next chapter! 👏🎓



October 2, 2025 at 11:28 AM
Congrats to Dr Michael Deistler @deismic.bsky.social, who defended his PhD!
Michael worked on "Machine Learning for Inference in Biophysical Neuroscience Simulations", focusing on simulation-based inference and differentiable simulation.
We wish him all the best for the next chapter! 👏🎓
Michael worked on "Machine Learning for Inference in Biophysical Neuroscience Simulations", focusing on simulation-based inference and differentiable simulation.
We wish him all the best for the next chapter! 👏🎓
The Macke lab is well-represented at the @bernsteinneuro.bsky.social conference in Frankfurt this year! We have lots of exciting new work to present with 7 posters (details👇) 1/9

September 30, 2025 at 2:06 PM
The Macke lab is well-represented at the @bernsteinneuro.bsky.social conference in Frankfurt this year! We have lots of exciting new work to present with 7 posters (details👇) 1/9
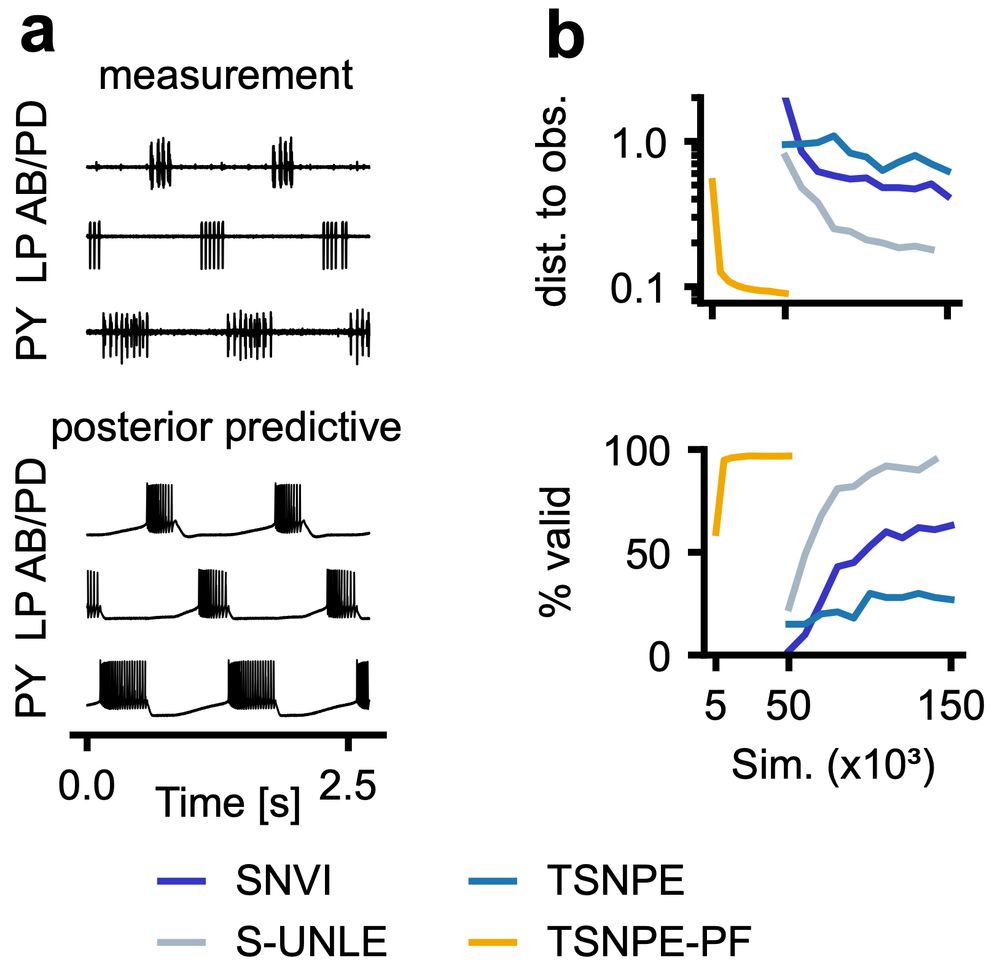
But does it scale to complex real-world problems? We tested it on two challenging Hodgkin-Huxley-type models:
🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
July 23, 2025 at 2:28 PM
But does it scale to complex real-world problems? We tested it on two challenging Hodgkin-Huxley-type models:
🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
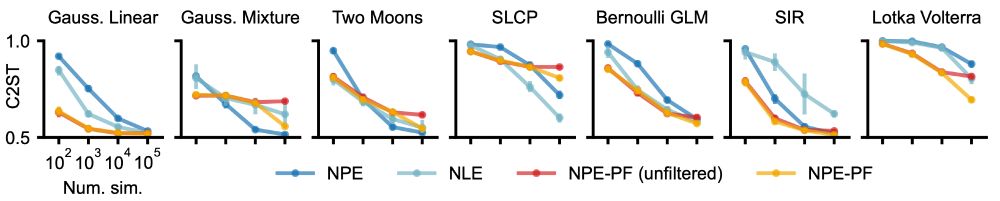
What you get with NPE-PF:
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
July 23, 2025 at 2:28 PM
What you get with NPE-PF:
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
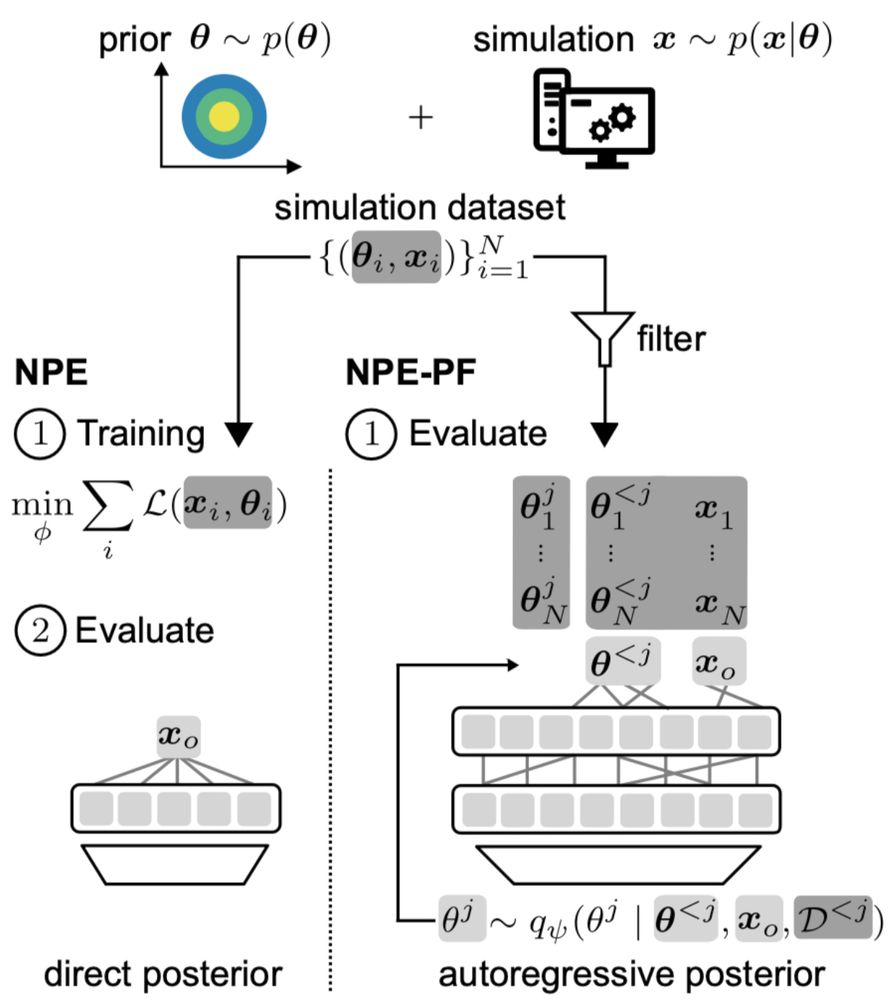
The key idea:
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
July 23, 2025 at 2:28 PM
The key idea:
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
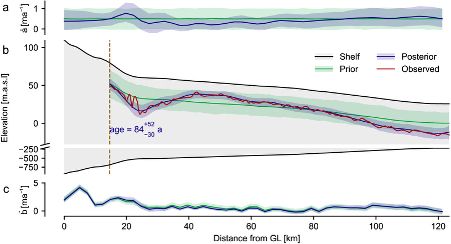
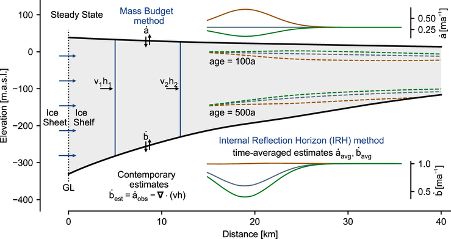
We obtain posterior distributions over ice accumulation and melting rates for Ekström Ice Shelf over the past hundreds of years. This allows us to make quantitative statements about the history of the atmospheric and oceanic conditions.
June 11, 2025 at 11:47 AM
We obtain posterior distributions over ice accumulation and melting rates for Ekström Ice Shelf over the past hundreds of years. This allows us to make quantitative statements about the history of the atmospheric and oceanic conditions.
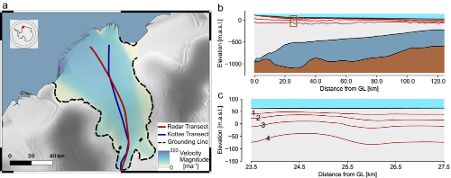
Thanks to great data collection efforts from @geophys-tuebingen.bsky.social and @awi.de, we can apply this approach to Ekström Ice Shelf, Antarctica.
June 11, 2025 at 11:47 AM
Thanks to great data collection efforts from @geophys-tuebingen.bsky.social and @awi.de, we can apply this approach to Ekström Ice Shelf, Antarctica.
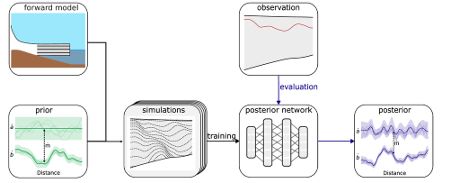
We develop a simulation-based-inference workflow for inferring the accumulation and melting rates from measurements of the internal layers.
June 11, 2025 at 11:47 AM
We develop a simulation-based-inference workflow for inferring the accumulation and melting rates from measurements of the internal layers.
Radar measurements have long been used for measuring the internal layer structure of Antarctic ice shelves. This structure contains information about the history of the ice shelf. This includes the past rate of snow accumulation at the surface, as well as ice melting at the base.
June 11, 2025 at 11:47 AM
Radar measurements have long been used for measuring the internal layer structure of Antarctic ice shelves. This structure contains information about the history of the ice shelf. This includes the past rate of snow accumulation at the surface, as well as ice melting at the base.
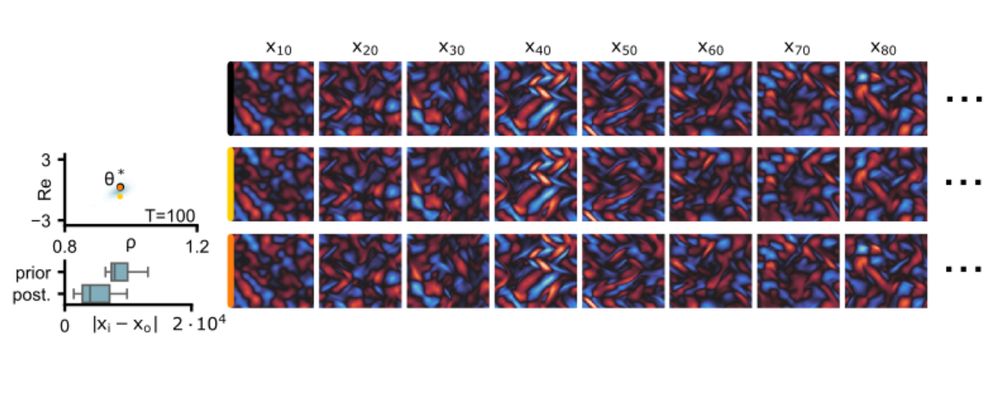
In FNSE, we only have to solve a smaller and easier inverse problem; it does scale relatively easily to high-dimensional simulators.
We validate this on a high-dimensional Kolmogorov flow simulator with around one million data dimensions.
We validate this on a high-dimensional Kolmogorov flow simulator with around one million data dimensions.
April 25, 2025 at 8:53 AM
In FNSE, we only have to solve a smaller and easier inverse problem; it does scale relatively easily to high-dimensional simulators.
We validate this on a high-dimensional Kolmogorov flow simulator with around one million data dimensions.
We validate this on a high-dimensional Kolmogorov flow simulator with around one million data dimensions.
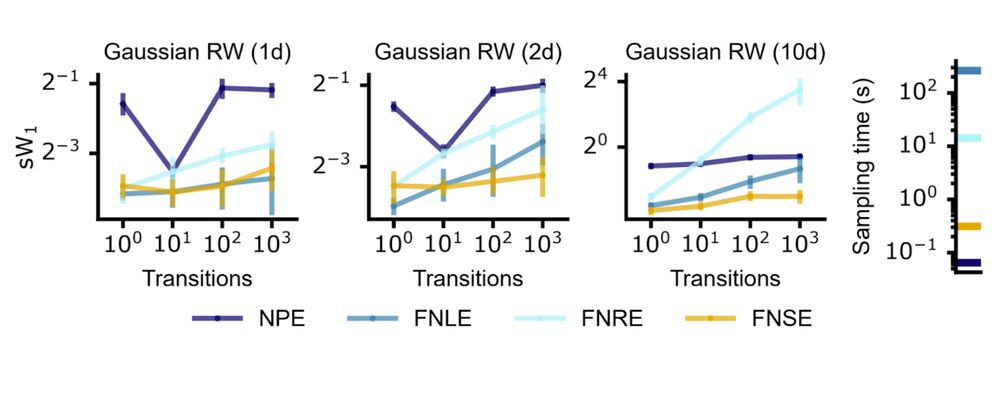
We apply this approach to various SBI methods (e.g. FNLE/FNRE), focusing on FNSE.
Compared to NPE with embedding nets, it’s more simulation-efficient and accurate across time series of varying lengths.
Compared to NPE with embedding nets, it’s more simulation-efficient and accurate across time series of varying lengths.
April 25, 2025 at 8:53 AM
We apply this approach to various SBI methods (e.g. FNLE/FNRE), focusing on FNSE.
Compared to NPE with embedding nets, it’s more simulation-efficient and accurate across time series of varying lengths.
Compared to NPE with embedding nets, it’s more simulation-efficient and accurate across time series of varying lengths.
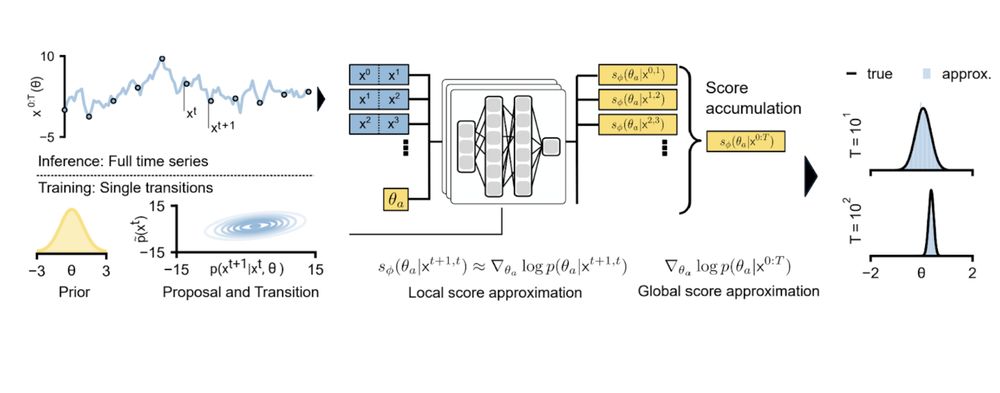
We propose an SBI approach that can exploit Markovian simulators by locally identifying parameters consistent with individual state transitions.
We then compose these local results to obtain a posterior over parameters that align with the entire time series observation.
We then compose these local results to obtain a posterior over parameters that align with the entire time series observation.
April 25, 2025 at 8:53 AM
We propose an SBI approach that can exploit Markovian simulators by locally identifying parameters consistent with individual state transitions.
We then compose these local results to obtain a posterior over parameters that align with the entire time series observation.
We then compose these local results to obtain a posterior over parameters that align with the entire time series observation.
Does any of this sound interesting to you? You might be excited to know that we are hiring PhD students and Postdocs!
Go to www.mackelab.org/jobs/ for details on more projects and how to contact us, or just find one of us (Auguste, Matthijs, Richard, Zina) at the conference—we're happy to chat!
Go to www.mackelab.org/jobs/ for details on more projects and how to contact us, or just find one of us (Auguste, Matthijs, Richard, Zina) at the conference—we're happy to chat!

March 27, 2025 at 2:03 PM
Does any of this sound interesting to you? You might be excited to know that we are hiring PhD students and Postdocs!
Go to www.mackelab.org/jobs/ for details on more projects and how to contact us, or just find one of us (Auguste, Matthijs, Richard, Zina) at the conference—we're happy to chat!
Go to www.mackelab.org/jobs/ for details on more projects and how to contact us, or just find one of us (Auguste, Matthijs, Richard, Zina) at the conference—we're happy to chat!
Finally, @rdgao.bsky.social is in another Tuesday workshop: “What biological details matter at mesoscopic scales?”
He will tell you about the good, the bad, and the ugly of getting more than what you had asked for using mechanistic models and probabilistic machine learning.
17:00, Corriveau/Sateux
He will tell you about the good, the bad, and the ugly of getting more than what you had asked for using mechanistic models and probabilistic machine learning.
17:00, Corriveau/Sateux

March 27, 2025 at 2:03 PM
Finally, @rdgao.bsky.social is in another Tuesday workshop: “What biological details matter at mesoscopic scales?”
He will tell you about the good, the bad, and the ugly of getting more than what you had asked for using mechanistic models and probabilistic machine learning.
17:00, Corriveau/Sateux
He will tell you about the good, the bad, and the ugly of getting more than what you had asked for using mechanistic models and probabilistic machine learning.
17:00, Corriveau/Sateux
We're also at the workshops!
Tuesday at 11:20, @auschulz.bsky.social will give a talk about deep generative models - VAEs and DDPMs - for linking neural activity and behavior, at the workshop on
"Building a foundation model for the brain" (Soutana 1).
Tuesday at 11:20, @auschulz.bsky.social will give a talk about deep generative models - VAEs and DDPMs - for linking neural activity and behavior, at the workshop on
"Building a foundation model for the brain" (Soutana 1).

March 27, 2025 at 2:03 PM
We're also at the workshops!
Tuesday at 11:20, @auschulz.bsky.social will give a talk about deep generative models - VAEs and DDPMs - for linking neural activity and behavior, at the workshop on
"Building a foundation model for the brain" (Soutana 1).
Tuesday at 11:20, @auschulz.bsky.social will give a talk about deep generative models - VAEs and DDPMs - for linking neural activity and behavior, at the workshop on
"Building a foundation model for the brain" (Soutana 1).
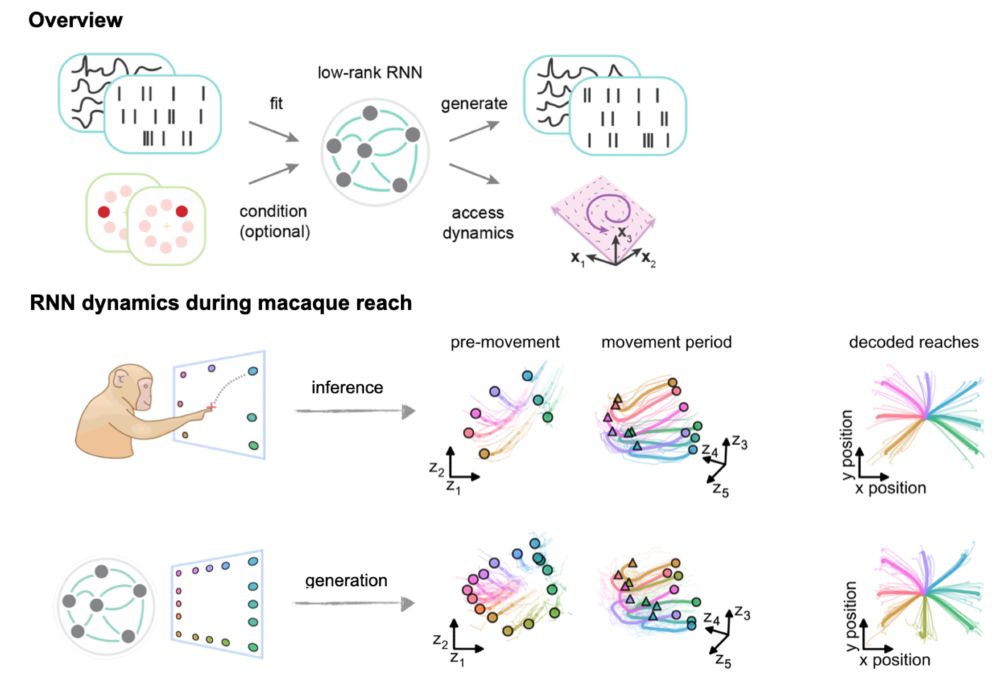
Interested in obtained models that are both interpretable and that can generate realistic neural data?
@matthijspals.bsky.social will give a contributed talk on fitting stochastic low-rank RNNs to neural data, Saturday 10:00 at the main conference! 🤩
@matthijspals.bsky.social will give a contributed talk on fitting stochastic low-rank RNNs to neural data, Saturday 10:00 at the main conference! 🤩
March 27, 2025 at 2:03 PM
Interested in obtained models that are both interpretable and that can generate realistic neural data?
@matthijspals.bsky.social will give a contributed talk on fitting stochastic low-rank RNNs to neural data, Saturday 10:00 at the main conference! 🤩
@matthijspals.bsky.social will give a contributed talk on fitting stochastic low-rank RNNs to neural data, Saturday 10:00 at the main conference! 🤩
In the same Friday session (Poster 2-039), @rdgao.bsky.social presents AutoMIND: using machine learning to infer parameters of spiking neural network models from brain recordings.
Come by if you hate fitting spiking networks or love degeneracy and invariances!
www.biorxiv.org/content/10.1...
Come by if you hate fitting spiking networks or love degeneracy and invariances!
www.biorxiv.org/content/10.1...

March 27, 2025 at 2:03 PM
In the same Friday session (Poster 2-039), @rdgao.bsky.social presents AutoMIND: using machine learning to infer parameters of spiking neural network models from brain recordings.
Come by if you hate fitting spiking networks or love degeneracy and invariances!
www.biorxiv.org/content/10.1...
Come by if you hate fitting spiking networks or love degeneracy and invariances!
www.biorxiv.org/content/10.1...
On Friday at 13:15 (Poster 2-012), Zina will talk about the role of recurrent connectivity for motion detection in the fruit fly.
Using connectome-constrained models of its visual system, we show how ablating recurrence decreases motion selectivity.
Using connectome-constrained models of its visual system, we show how ablating recurrence decreases motion selectivity.

March 27, 2025 at 2:03 PM
On Friday at 13:15 (Poster 2-012), Zina will talk about the role of recurrent connectivity for motion detection in the fruit fly.
Using connectome-constrained models of its visual system, we show how ablating recurrence decreases motion selectivity.
Using connectome-constrained models of its visual system, we show how ablating recurrence decreases motion selectivity.
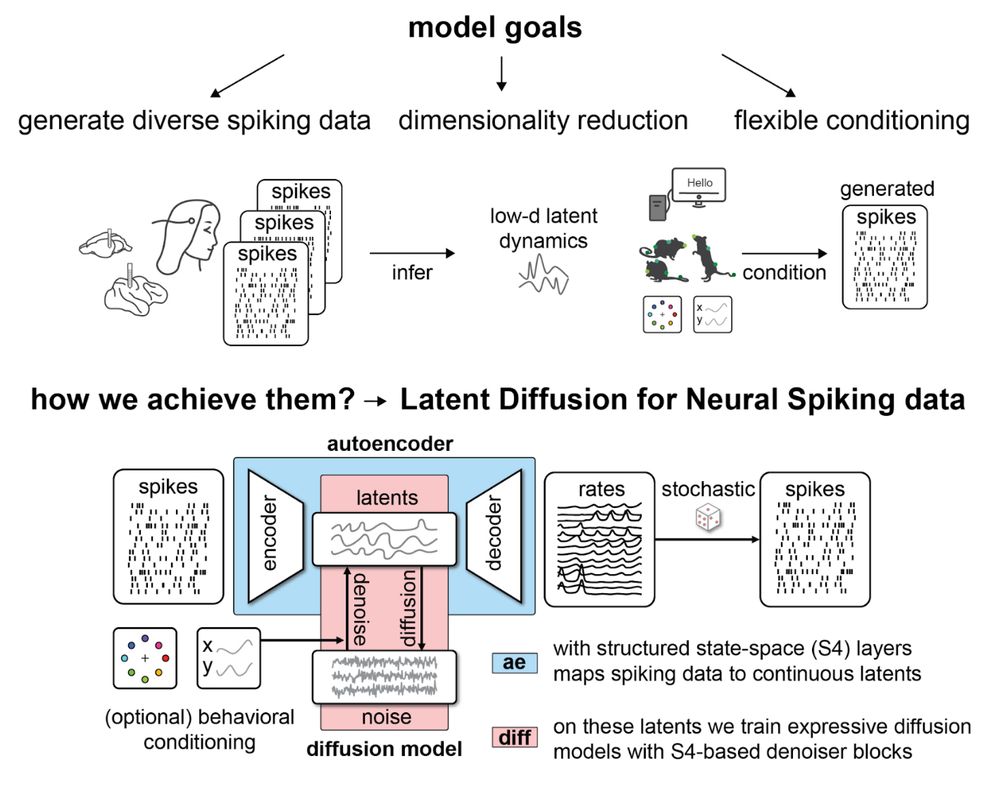
On Thursday at 20:00 (Poster 1-108), @auschulz.bsky.social introduces LDNS, a diffusion-based latent variable model to generate diverse neural spiking data flexibly conditioned on external variables.

March 27, 2025 at 2:03 PM
On Thursday at 20:00 (Poster 1-108), @auschulz.bsky.social introduces LDNS, a diffusion-based latent variable model to generate diverse neural spiking data flexibly conditioned on external variables.
The @mackelab.bsky.social is represented at @cosynemeeting.bsky.social #cosyne2025 in Montreal
with 3 posters, 2 workshop talks, and a main conference contributed talk (for the very first time in Mackelab history 🎉)!
with 3 posters, 2 workshop talks, and a main conference contributed talk (for the very first time in Mackelab history 🎉)!

March 27, 2025 at 2:03 PM
The @mackelab.bsky.social is represented at @cosynemeeting.bsky.social #cosyne2025 in Montreal
with 3 posters, 2 workshop talks, and a main conference contributed talk (for the very first time in Mackelab history 🎉)!
with 3 posters, 2 workshop talks, and a main conference contributed talk (for the very first time in Mackelab history 🎉)!
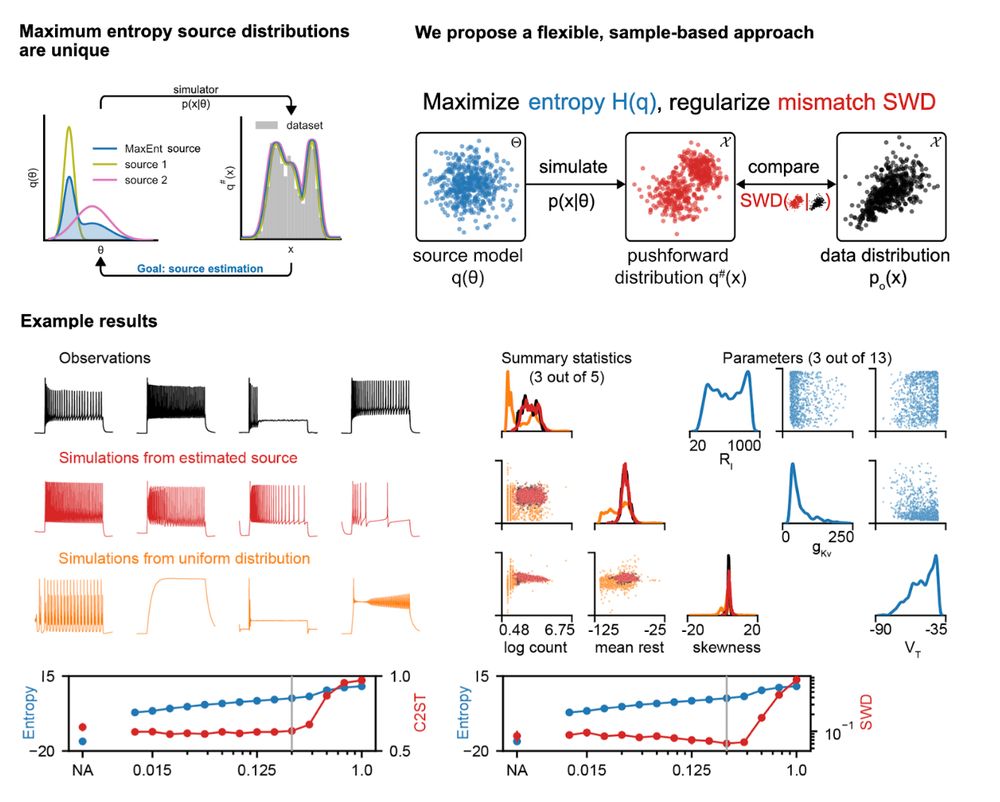
We introduce a new maximum-entropy, sample-based approach to solve the source distribution estimation problem. Poster #4006 (East; Fri 13 Dec 11PT). By @vetterj.bsky.social, @gmoss13.bsky.social ➡️ openreview.net/forum?id=0cg... 4/4
December 9, 2024 at 7:28 PM
We introduce a new maximum-entropy, sample-based approach to solve the source distribution estimation problem. Poster #4006 (East; Fri 13 Dec 11PT). By @vetterj.bsky.social, @gmoss13.bsky.social ➡️ openreview.net/forum?id=0cg... 4/4
@jkapoor.bsky.social and @auschulz.bsky.social introduce LDNS, a diffusion-based latent variable model to generate diverse neural spiking data flexibly conditioned on external variables Poster #4010 (East; Wed 11 Dec 11PT) ➡️ openreview.net/forum?id=ZX6... 3/4
December 9, 2024 at 7:28 PM
@jkapoor.bsky.social and @auschulz.bsky.social introduce LDNS, a diffusion-based latent variable model to generate diverse neural spiking data flexibly conditioned on external variables Poster #4010 (East; Wed 11 Dec 11PT) ➡️ openreview.net/forum?id=ZX6... 3/4
We show how to generate long sequences of realistic neural data with stochastic low-rank RNNs - and how to find their fixed points. Poster #3908 (East; Wed 11 Dec 11PT). @matthijspals.bsky.social, @aesagtekin.bsky.social, F Pei, M Gloeckler, @jakhmack.bsky.social ➡️ openreview.net/forum?id=C0E... 2/4

December 9, 2024 at 7:28 PM
We show how to generate long sequences of realistic neural data with stochastic low-rank RNNs - and how to find their fixed points. Poster #3908 (East; Wed 11 Dec 11PT). @matthijspals.bsky.social, @aesagtekin.bsky.social, F Pei, M Gloeckler, @jakhmack.bsky.social ➡️ openreview.net/forum?id=C0E... 2/4
Hi world! This is the brand-new BlueSky account of the Machine Learning in Science (@jakhmack.bsky.social) lab. We create probabilistic #MachineLearning and #AI Tools for scientific discovery — but more on that soon!
For now let’s introduce ourselves with some pictures of our recent group retreat.
For now let’s introduce ourselves with some pictures of our recent group retreat.


November 14, 2024 at 1:39 PM
Hi world! This is the brand-new BlueSky account of the Machine Learning in Science (@jakhmack.bsky.social) lab. We create probabilistic #MachineLearning and #AI Tools for scientific discovery — but more on that soon!
For now let’s introduce ourselves with some pictures of our recent group retreat.
For now let’s introduce ourselves with some pictures of our recent group retreat.

