
Host-Defence | Structural biology | Single-molecule Biophysics
http://www.loefflab.com





reveals that the PYROXD1 CTD engages with the
catalytic center cleft of RTCB, thereby blocking the
substrate binding sites and rendering the catalytic
center inaccessible. Mutation of the interacting residues resulted in a loss of protection.

reveals that the PYROXD1 CTD engages with the
catalytic center cleft of RTCB, thereby blocking the
substrate binding sites and rendering the catalytic
center inaccessible. Mutation of the interacting residues resulted in a loss of protection.
















Today in @cellpress.bsky.social we report the structure and function of the Shedu anti-phage defense system.
tinyurl.com/4crj6dnx
A long 🧵...
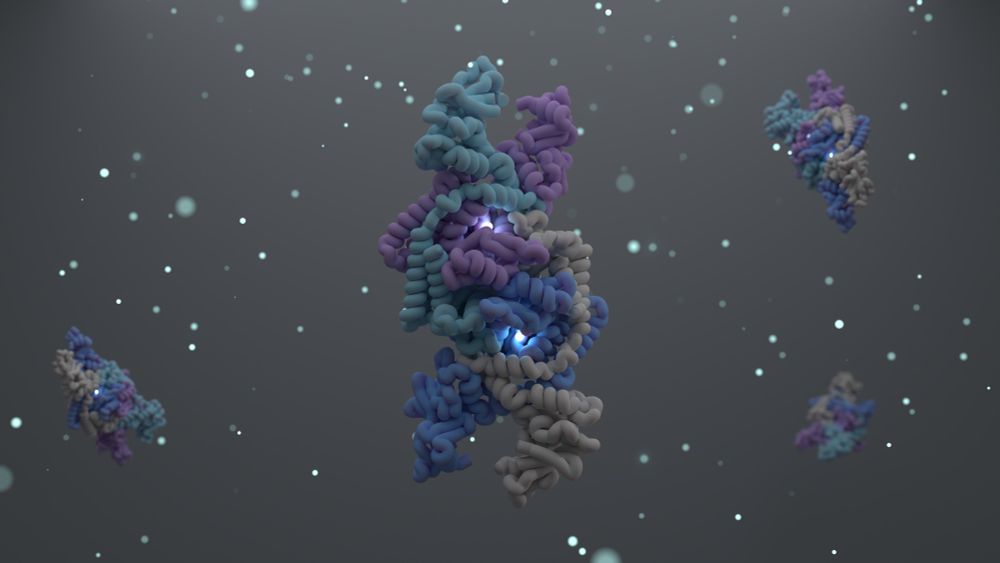
Today in @cellpress.bsky.social we report the structure and function of the Shedu anti-phage defense system.
tinyurl.com/4crj6dnx
A long 🧵...
Earlier this year we showed how the DdmDE system from pandemic Vibrio cholerae defends against invading plasmids. A great collaboration between the labs of Martin Jinek (UZH) and @mblokesch.bsky.social (EPFL).
www.science.org/doi/10.1126/...

Earlier this year we showed how the DdmDE system from pandemic Vibrio cholerae defends against invading plasmids. A great collaboration between the labs of Martin Jinek (UZH) and @mblokesch.bsky.social (EPFL).
www.science.org/doi/10.1126/...

