
http://lcolladotor.github.io
You'll be able to listen to it next Friday or so at www.iheart.com/podcast/269-.... Thanks Clau from iHeartRadio Miami's "Estamos Contigo" podcast for reaching out!
I'll be talking in Spanish about our recent #HabenulaLIBD 📜
bsky.app/profile/lcol...

You'll be able to listen to it next Friday or so at www.iheart.com/podcast/269-.... Thanks Clau from iHeartRadio Miami's "Estamos Contigo" podcast for reaching out!
I'll be talking in Spanish about our recent #HabenulaLIBD 📜
bsky.app/profile/lcol...









Feel free to come visit us in Baltimore 🦀 whenever you want ^^

Feel free to come visit us in Baltimore 🦀 whenever you want ^^
Thanks @jef.works for inviting me! And thanks @mikeschatz.bsky.social @jihk.bsky.social for your guidance! Great to share this experience with Fred Tan as well ^_^
It’s exciting to talk about […]

Thanks @jef.works for inviting me! And thanks @mikeschatz.bsky.social @jihk.bsky.social for your guidance! Great to share this experience with Fred Tan as well ^_^
It’s exciting to talk about […]
@isaacgs94.bsky.social: Are you Julio Collado?
Leo: no
I: 😥
L: I’m his son
I:
L: I’m Leo Collado
Isaac had seen me on social media. We already followed each other here 😅
Congrats on completing your PhD studies at @sangerinstitute.bsky.social Enjoy the celebrations! 🇲🇽

@isaacgs94.bsky.social: Are you Julio Collado?
Leo: no
I: 😥
L: I’m his son
I:
L: I’m Leo Collado
Isaac had seen me on social media. We already followed each other here 😅
Congrats on completing your PhD studies at @sangerinstitute.bsky.social Enjoy the celebrations! 🇲🇽
lcolladotor.github.io/2025/03/14/i...
I describe our use case and why I think that Posit Connect will be helpful for our @lieberinstitute.bsky.social work moving forward
@posit.co #RStats #shiny #shinyapps #spatialLIBD #iSEE

lcolladotor.github.io/2025/03/14/i...
I describe our use case and why I think that Posit Connect will be helpful for our @lieberinstitute.bsky.social work moving forward
@posit.co #RStats #shiny #shinyapps #spatialLIBD #iSEE
youtu.be/alL-g2roXxM
We go over what analyses our @lieberinstitute.bsky.social has expertise on 💻
Slides orchestrated by @lahuuki.bsky.social at speakerdeck.com/lahuuki/2025...
👥 go.bsky.app/53gy4UP


youtu.be/alL-g2roXxM
We go over what analyses our @lieberinstitute.bsky.social has expertise on 💻
Slides orchestrated by @lahuuki.bsky.social at speakerdeck.com/lahuuki/2025...
👥 go.bsky.app/53gy4UP
It's great to see current students re-implement the Cultural Mixers @mandymejia.bsky.social & I ran in 2012-~2015 lcolladotor.github.io/2013/11/20/s...
📸 with @mgschiavon.bsky.social @surh.bsky.social

It's great to see current students re-implement the Cultural Mixers @mandymejia.bsky.social & I ran in 2012-~2015 lcolladotor.github.io/2013/11/20/s...
📸 with @mgschiavon.bsky.social @surh.bsky.social
* RStudio project +
* Git repo +
* GitHub website using the #postcards #rstats 📦 by @seankross.com
It's been great to be part of your academic journey!
lcolladotor.github.io/rnaseq_LCG-U...


* RStudio project +
* Git repo +
* GitHub website using the #postcards #rstats 📦 by @seankross.com
It's been great to be part of your academic journey!
lcolladotor.github.io/rnaseq_LCG-U...
“you may want to arrange your travel plans—especially if travel is nonessential—to return to the U.S. no later than January 19.”
@johnshopkinssph.bsky.social
ois.jhu.edu/travel-infor...
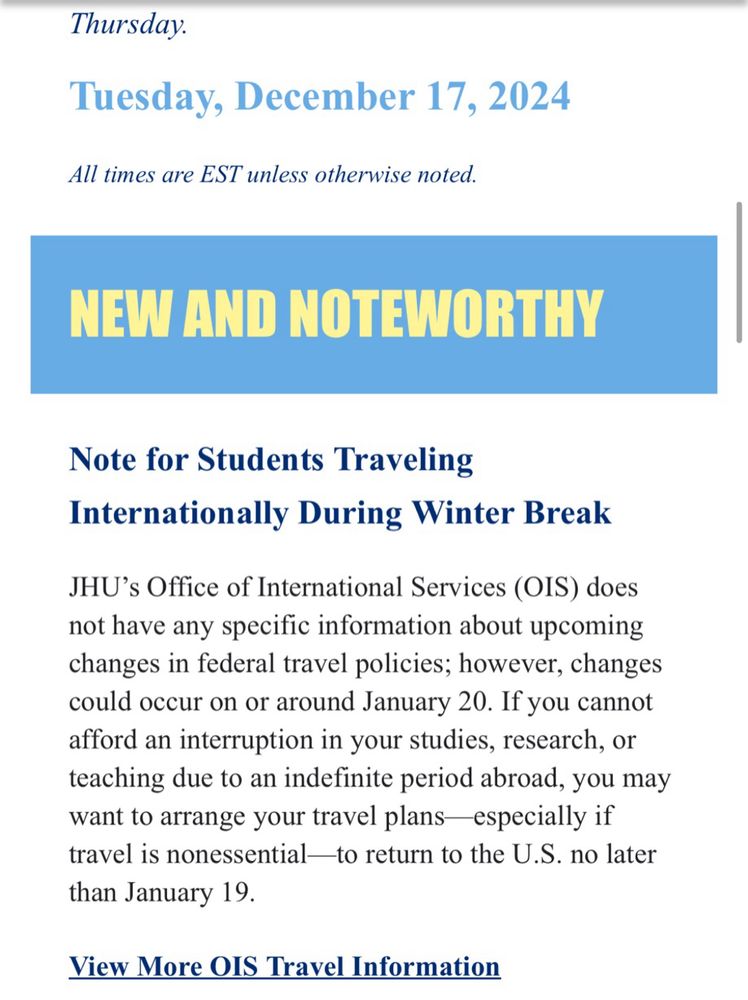
“you may want to arrange your travel plans—especially if travel is nonessential—to return to the U.S. no later than January 19.”
@johnshopkinssph.bsky.social
ois.jhu.edu/travel-infor...
Although github-readme-stats.vercel.app/api/top-lang... says that I mostly write html code :P

Although github-readme-stats.vercel.app/api/top-lang... says that I mostly write html code :P
raw.githubusercontent.com/LiNk-NY/LiNK... --> github.com/lcolladotor/...
raw.githubusercontent.com/LiNk-NY/LiNK... --> github.com/lcolladotor/...
We’ve also used C2L with @lieberinstitute.bsky.social data
#SpatialBiology

We’ve also used C2L with @lieberinstitute.bsky.social data
#SpatialBiology
Using Visium, Xenium, and Hyperion data

Using Visium, Xenium, and Hyperion data
The image below is from a previous slide showing how the results from #CellNEST can be visualized 🎨
I’m looking forward to the paper 📜!
#SpatialBiology
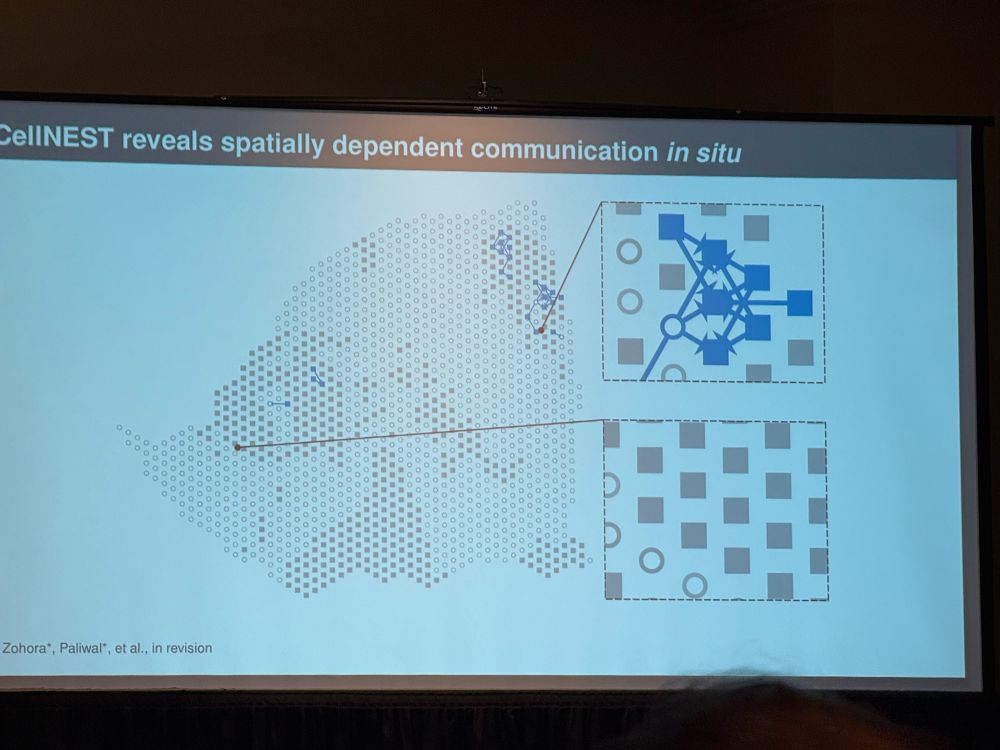
The image below is from a previous slide showing how the results from #CellNEST can be visualized 🎨
I’m looking forward to the paper 📜!
#SpatialBiology
That’s great to see! It’s hard to validate cell to cell communication results 😣
#SpatialBiology

That’s great to see! It’s hard to validate cell to cell communication results 😣
#SpatialBiology


