Than you are more than welcome to apply to join my group starting Jan 2026 :)
candidate.hr-manager.net/ApplicationI...
Please reach out if you have any questions!

Than you are more than welcome to apply to join my group starting Jan 2026 :)
candidate.hr-manager.net/ApplicationI...
Please reach out if you have any questions!

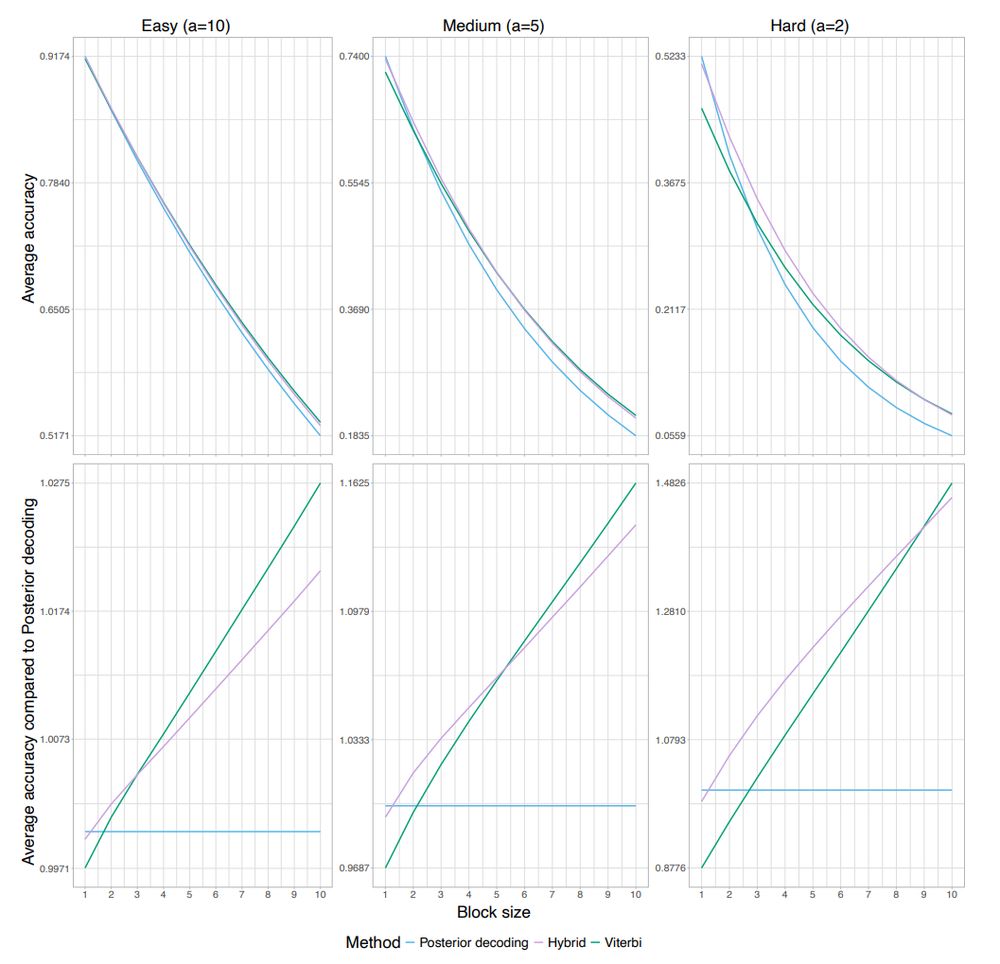

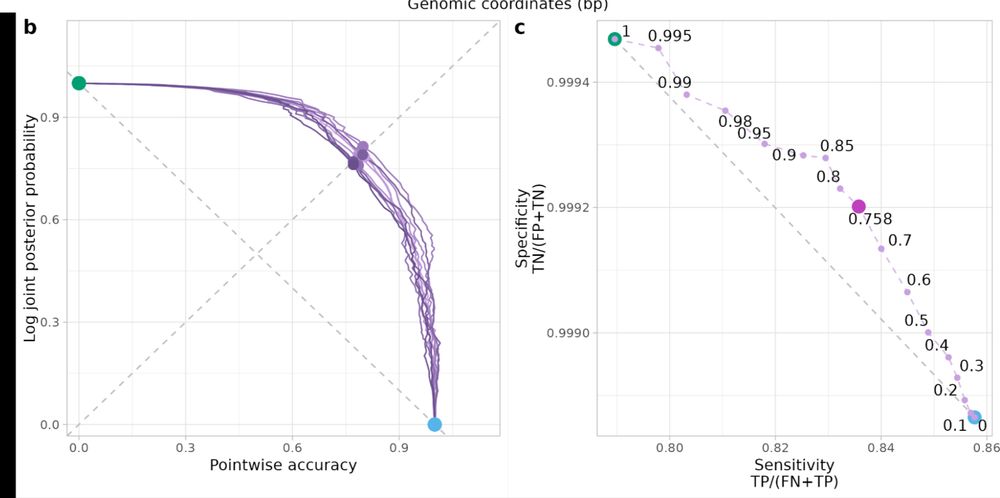

arxiv.org/pdf/2504.15156
www.biorxiv.org/content/10.1...

arxiv.org/pdf/2504.15156
www.biorxiv.org/content/10.1...





Link for application:
candidate.hr-manager.net/ApplicationI...

Link for application:
candidate.hr-manager.net/ApplicationI...

