Vanni Lab at UNIFR, Switzerland
@labvanni.bsky.social
Computational biophysics and other amenities...
"More thinking and less pipetting"
"More thinking and less pipetting"
Well...for once I can repost/like a paper I read very carefully 😆
August 21, 2025 at 1:07 PM
Well...for once I can repost/like a paper I read very carefully 😆
Congratulations André and team. Very very nice work!
August 21, 2025 at 11:26 AM
Congratulations André and team. Very very nice work!
All trajectories and in Zenodo, get in contact if you have questions!
August 7, 2025 at 8:45 AM
All trajectories and in Zenodo, get in contact if you have questions!
Of course! all trajectories are in Zenodo: doi.org/10.5281/zeno...
If you want more details, just get in touch!
If you want more details, just get in touch!
A conserved mechanism of membrane fusion in nuclear pore complex assembly
Molecular dynamics (MD) simulation data for the preprint titled "A conserved mechanism of membrane fusion in nuclear pore complex assembly" are organized into six folders, each named according to the ...
doi.org
July 25, 2025 at 3:30 PM
Of course! all trajectories are in Zenodo: doi.org/10.5281/zeno...
If you want more details, just get in touch!
If you want more details, just get in touch!
Altogether, we think that the mechanism of membrane fusion described in the context of NPC assembly here is different from what known for other fusion machineries (e.g. SNAREs, dynamin-like GTPases,…). More details in the manuscript—let us know your thoughts! (13/13)
July 23, 2025 at 11:55 AM
Altogether, we think that the mechanism of membrane fusion described in the context of NPC assembly here is different from what known for other fusion machineries (e.g. SNAREs, dynamin-like GTPases,…). More details in the manuscript—let us know your thoughts! (13/13)
Finally, phylogenetic analyses showed that Brl1/Brr6 homologues are broadly distributed across eukaryotes, and functional experiments in human cells and fly establish CLCC1 as an NPC fusogen in metazoans, consistent with recent preprints by @olzmannlab.bsky.social, Xiao-Wei Chen and others. (12/13)
July 23, 2025 at 11:54 AM
Finally, phylogenetic analyses showed that Brl1/Brr6 homologues are broadly distributed across eukaryotes, and functional experiments in human cells and fly establish CLCC1 as an NPC fusogen in metazoans, consistent with recent preprints by @olzmannlab.bsky.social, Xiao-Wei Chen and others. (12/13)
We also modelled Brl1 in curved membranes (a vesicle mimicking the INM shape). Same result: Brl1/Brr6 oligomers stably interacting and promoting lipid mixing within the channel. (11/13)
July 23, 2025 at 11:53 AM
We also modelled Brl1 in curved membranes (a vesicle mimicking the INM shape). Same result: Brl1/Brr6 oligomers stably interacting and promoting lipid mixing within the channel. (11/13)
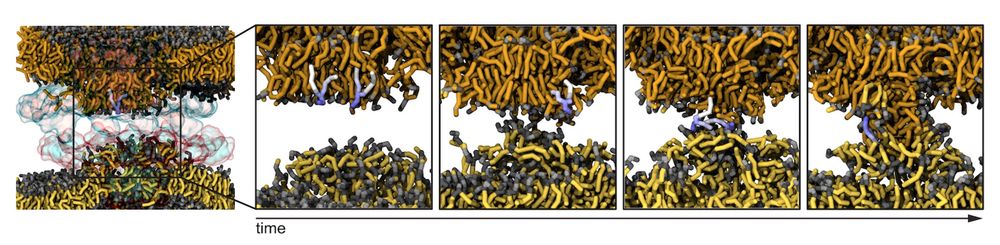
When observed closely, lipid mixing looks like classic hemifusion-like intermediates with exposed lipid-tails (but stabilized within protein rings here). (10/13)
July 23, 2025 at 11:51 AM
When observed closely, lipid mixing looks like classic hemifusion-like intermediates with exposed lipid-tails (but stabilized within protein rings here). (10/13)
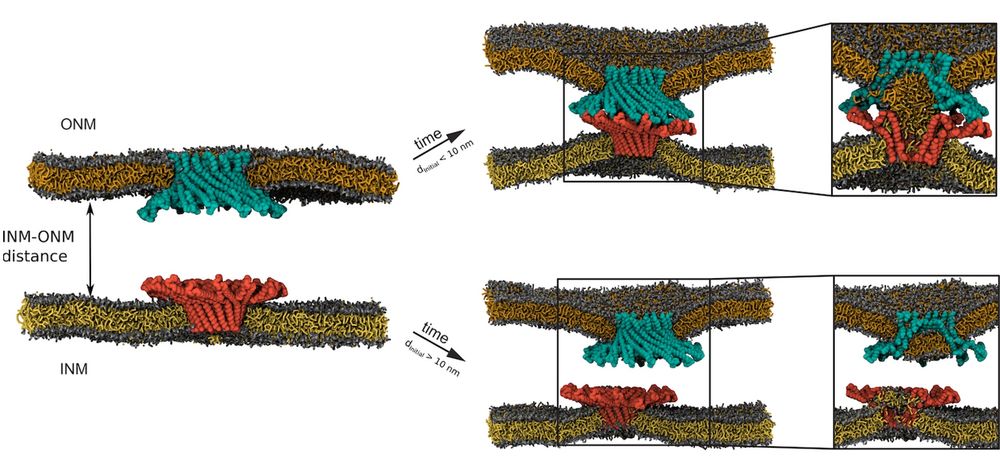
But here is another interesting part: interaction between Brl1 and Brr6 only happens when the membranes are < 10 nm apart. This is also very consistent with previous measurement of these distances in NPC assembly intermediates prior to fusion. (9/13)
July 23, 2025 at 11:51 AM
But here is another interesting part: interaction between Brl1 and Brr6 only happens when the membranes are < 10 nm apart. This is also very consistent with previous measurement of these distances in NPC assembly intermediates prior to fusion. (9/13)
We went back to simulations of opposed bilayers, but with protein oligomers in both of them. We observed a gradual decrease in membrane distances to the point of interaction between proteins (Brl1 and Brr6). This interaction results in lipid mixing in the central channel of the rings. (8/13)
July 23, 2025 at 11:50 AM
We went back to simulations of opposed bilayers, but with protein oligomers in both of them. We observed a gradual decrease in membrane distances to the point of interaction between proteins (Brl1 and Brr6). This interaction results in lipid mixing in the central channel of the rings. (8/13)
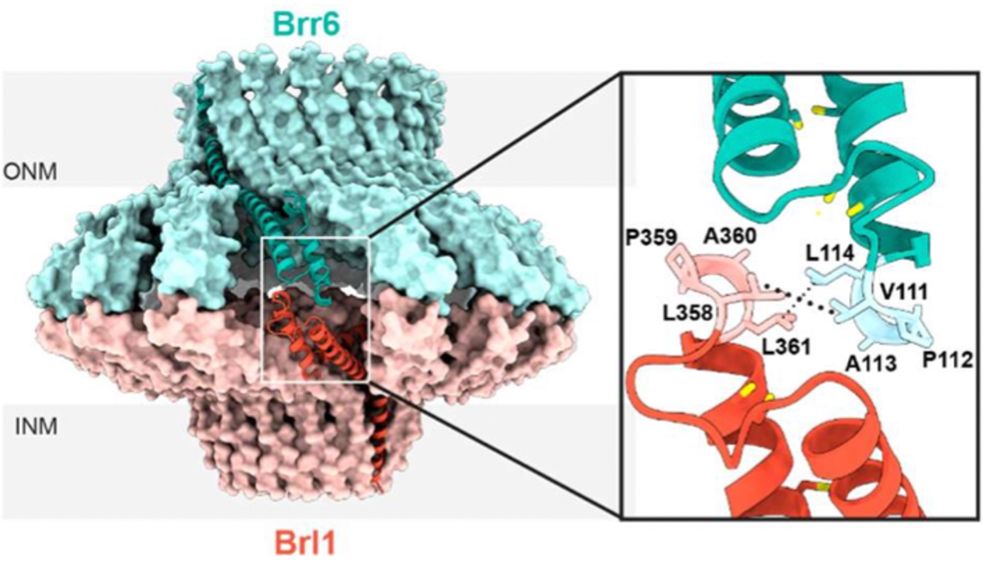
This hydrophobic loop motif is highly conserved in fungi, and disrupting this loop halted NPC assembly process. This is telling us that Brl1/Brr6 head-to-head interaction is essential for NPC assembly. (7/13)
July 23, 2025 at 11:48 AM
This hydrophobic loop motif is highly conserved in fungi, and disrupting this loop halted NPC assembly process. This is telling us that Brl1/Brr6 head-to-head interaction is essential for NPC assembly. (7/13)
Given that both Brl1 and Brr6 are essential, and their interaction has been studied in earlier work, we modeled 16 copies of Brl1 together with 16 of Brr6. We got a 32-mer complex with two subunits of 16-mers of Brl1/Brr6 with a head-to-head interaction mediated by a hydrophobic loop motif. (6/13)
July 23, 2025 at 11:48 AM
Given that both Brl1 and Brr6 are essential, and their interaction has been studied in earlier work, we modeled 16 copies of Brl1 together with 16 of Brr6. We got a 32-mer complex with two subunits of 16-mers of Brl1/Brr6 with a head-to-head interaction mediated by a hydrophobic loop motif. (6/13)

