rdcu.be/ezGqT
Work led by @sobonnal.bsky.social, with Simon Bajew & @rmartinezcorral.bsky.social. @crg.eu @upf.edu

rdcu.be/ezGqT
Work led by @sobonnal.bsky.social, with Simon Bajew & @rmartinezcorral.bsky.social. @crg.eu @upf.edu
Please RT!
We're looking for a postdoc to join an exciting joint project between our lab @upf.edu & @crg.eu (Barcelona) and the Sander lab @mdc-berlin.bsky.social (Berlin) investigating how alternative splicing and microexons influences the maturation of pancreatic islets.
Deadline: 30/09/25👇
Please RT!
We're looking for a postdoc to join an exciting joint project between our lab @upf.edu & @crg.eu (Barcelona) and the Sander lab @mdc-berlin.bsky.social (Berlin) investigating how alternative splicing and microexons influences the maturation of pancreatic islets.
Deadline: 30/09/25👇
www.sciencedirect.com/science/arti...
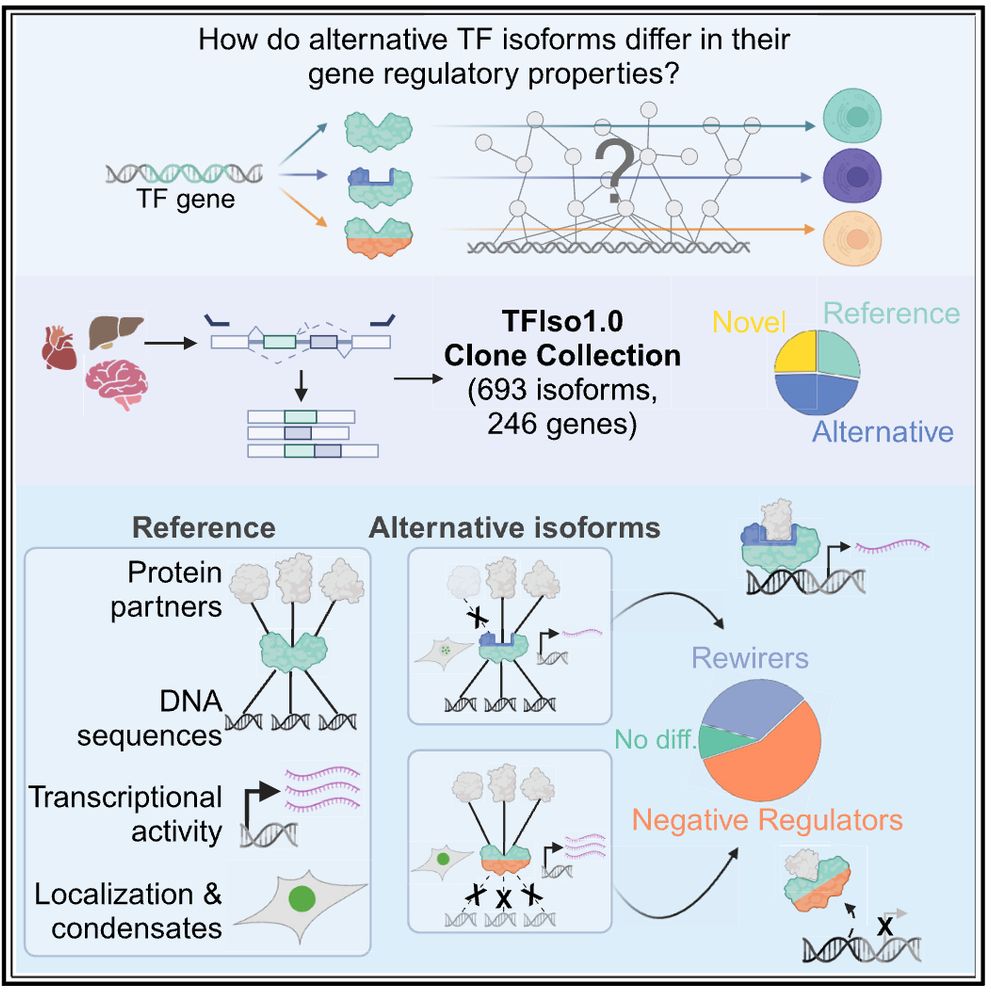
we cloned 100's of isoforms (that arise through alternative splicing etc.) testing them in high-throughput assays for DNA-binding, protein-binding, activation, and more.
www.sciencedirect.com/science/arti...
we cloned 100's of isoforms (that arise through alternative splicing etc.) testing them in high-throughput assays for DNA-binding, protein-binding, activation, and more.
In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp
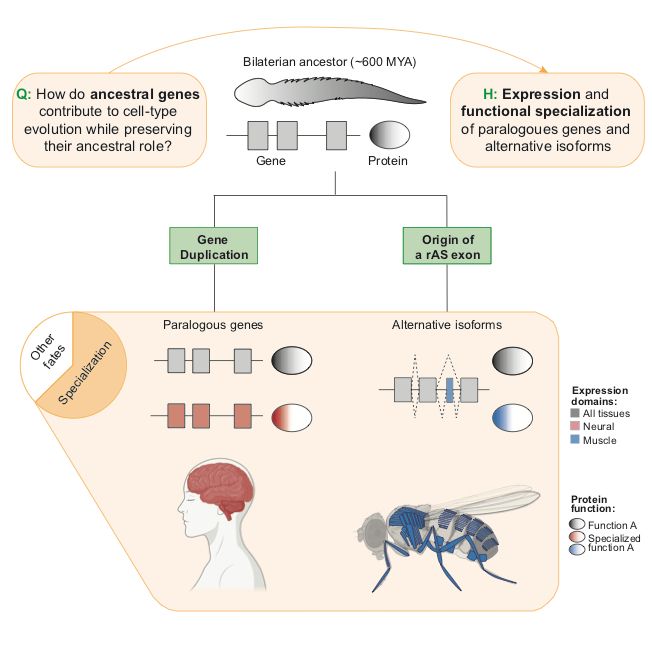
In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp

