
ERC Research, FensKavli Scholar.
Molecular basis of neuroplasticity.
New genomics technologies.
https://www.kitazawa-lab.com/home
Our lab is now applying HisTrac to study id specification and plasticity (e.g., memory) and we look forward to sharing exciting results soon!

Our lab is now applying HisTrac to study id specification and plasticity (e.g., memory) and we look forward to sharing exciting results soon!
Indeed, your nanobody-Tn5 protocol is AMAZING!
We actually put some modifications, like sample-bcd implementation, as well as replacement of enzyme for 10x reaction, which may be interesting for you.
I look forward to discussing with you again!
Indeed, your nanobody-Tn5 protocol is AMAZING!
We actually put some modifications, like sample-bcd implementation, as well as replacement of enzyme for 10x reaction, which may be interesting for you.
I look forward to discussing with you again!
Our lab is now applying HisTrac to study id specification and plasticity (e.g., memory) and we look forward to sharing exciting results soon!

Our lab is now applying HisTrac to study id specification and plasticity (e.g., memory) and we look forward to sharing exciting results soon!

Using scHisTrac-seq to profile transcriptome, chromatin accessibility, and bivalency during neurodifferentiation, we analyzed over 90,000 single cells and uncovered drastic cell identity transitions—namely “identity jumps.

Using scHisTrac-seq to profile transcriptome, chromatin accessibility, and bivalency during neurodifferentiation, we analyzed over 90,000 single cells and uncovered drastic cell identity transitions—namely “identity jumps.
For the final form, we developed scHisTrac-seq, enabling simultaneous profiling of past and present multiomic states from the same single cells via a nanobody-mediated dual scCUT&Tag protocol (past = m6A, present = H3K27ac, transcription marker).
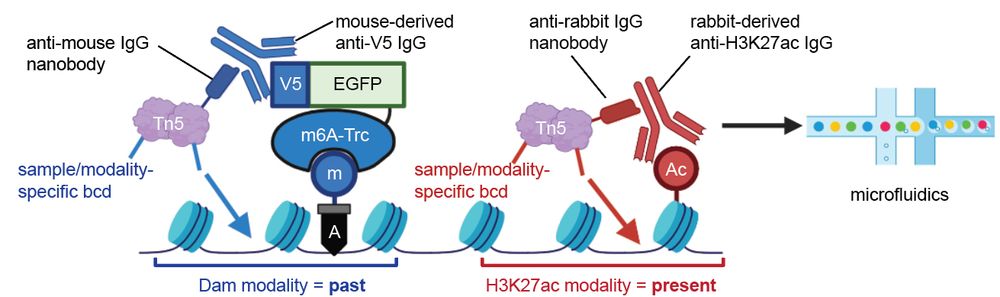
For the final form, we developed scHisTrac-seq, enabling simultaneous profiling of past and present multiomic states from the same single cells via a nanobody-mediated dual scCUT&Tag protocol (past = m6A, present = H3K27ac, transcription marker).

This captured neonatal epigenome and transcriptome in the adult brain, demonstrating that HisTrac-seq remains effective for at least two months in vivo!

This captured neonatal epigenome and transcriptome in the adult brain, demonstrating that HisTrac-seq remains effective for at least two months in vivo!
In addition, we extended the validation to the mesodermal cardiomyocyte lineage.

In addition, we extended the validation to the mesodermal cardiomyocyte lineage.

Because previously shown that m6A can persist for several hours until the next DNA replication, we expected it to serve as a long-term ”bookmark” in neurons, potentially for months.

Because previously shown that m6A can persist for several hours until the next DNA replication, we expected it to serve as a long-term ”bookmark” in neurons, potentially for months.

Following threads will elaborate on HisTrac-seq and and our findings.

Following threads will elaborate on HisTrac-seq and and our findings.

