
She/Her :) A science, dog, photography person, and most of all, an absolute chromatin geek

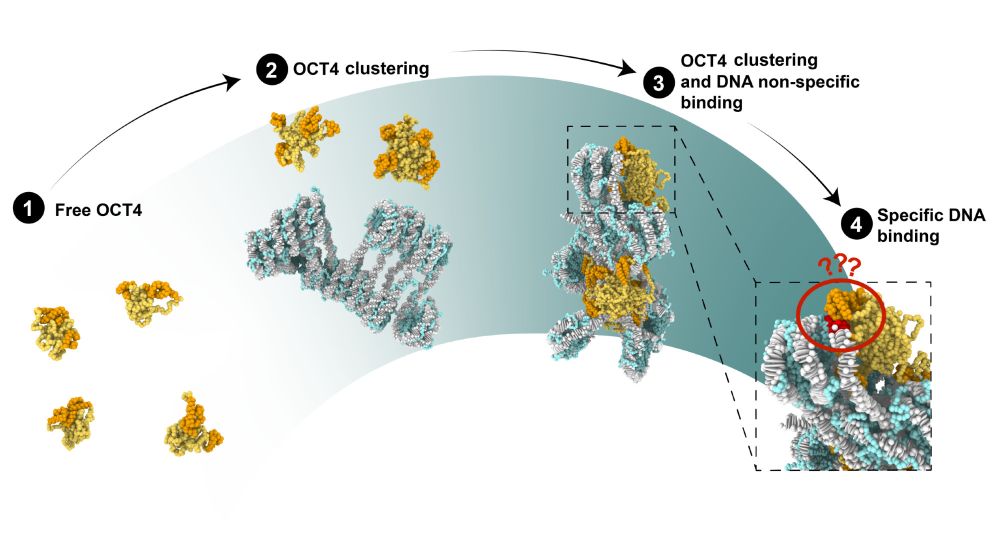
Very happy and proud of this Rosen/Redding/Collepardo collaborative effort 🔥
also on bky:
@rcollepardo.bsky.social @janhuemar.bsky.social
Very happy and proud of this Rosen/Redding/Collepardo collaborative effort 🔥
also on bky:
@rcollepardo.bsky.social @janhuemar.bsky.social


The key metric? Inter-fiber valency (how many neighbours each fiber contacts).
Higher valency in arrays (namely, arrays close to 10N+5 linkers) make for stronger inter-fiber interactions, and more stable condensates.

The key metric? Inter-fiber valency (how many neighbours each fiber contacts).
Higher valency in arrays (namely, arrays close to 10N+5 linkers) make for stronger inter-fiber interactions, and more stable condensates.
We found that even subtle changes in spacing lead to large-scale structural differences (!)
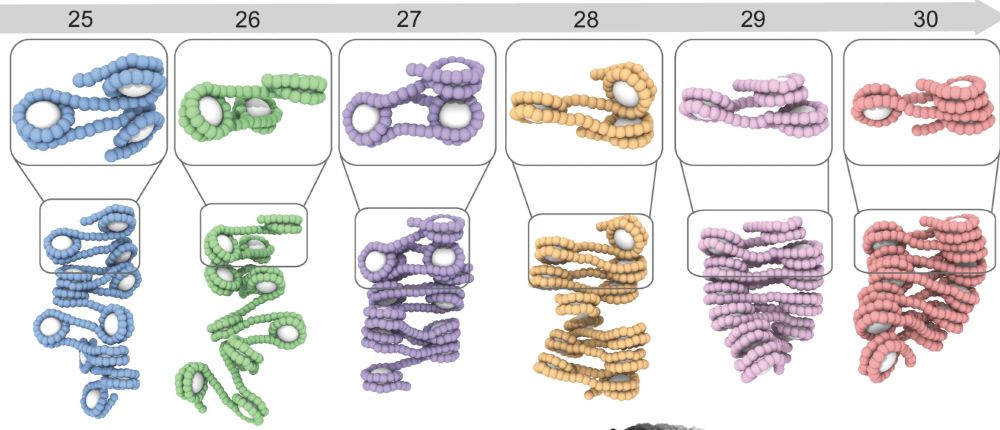
We found that even subtle changes in spacing lead to large-scale structural differences (!)
Spacing between them matters: Just changing the DNA length between nucleosomes, even by a single base pair, dramatically alters how they interact and phase separate.


Spacing between them matters: Just changing the DNA length between nucleosomes, even by a single base pair, dramatically alters how they interact and phase separate.

