
Javier Santoyo
@jsantoyo.bsky.social
Genomics & Bioinformatics Scientist at the University of Edinburgh | Head of Edinburgh Genomics | #Omics #NGS #LongReads 🧪🧬 🖥️
Opinions my own. He/Him
Lab: https://genomics.ed.ac.uk/
LinkedIn: https://www.linkedin.com/in/javiersantoyolopez/
Opinions my own. He/Him
Lab: https://genomics.ed.ac.uk/
LinkedIn: https://www.linkedin.com/in/javiersantoyolopez/
insilicoSV: a flexible grammar-based framework for structural variant simulation and placement. #StructuralVariants #SVs #SVsimulation #SimulatedData #Genomics #Bioinformatics 🧪🧬 🖥️
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...

November 9, 2025 at 8:05 PM
insilicoSV: a flexible grammar-based framework for structural variant simulation and placement. #StructuralVariants #SVs #SVsimulation #SimulatedData #Genomics #Bioinformatics 🧪🧬 🖥️
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...
Genome size estimation from long read overlaps. #GenomeSize #LongRead #Sequencing #Genomics #Bioinformatics 🧪🧬 🖥️
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...

November 9, 2025 at 7:05 PM
Genome size estimation from long read overlaps. #GenomeSize #LongRead #Sequencing #Genomics #Bioinformatics 🧪🧬 🖥️
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...
HI-FEVER: a Nextflow pipeline for the high-throughput discovery and annotation of endogenous viral elements. #EndogenousViralElements #EukaryoticHostGenomes #Genomics #Bioinformatics 🧪🧬 🖥️
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...

November 9, 2025 at 6:05 PM
HI-FEVER: a Nextflow pipeline for the high-throughput discovery and annotation of endogenous viral elements. #EndogenousViralElements #EukaryoticHostGenomes #Genomics #Bioinformatics 🧪🧬 🖥️
academic.oup.com/bioinformati...
academic.oup.com/bioinformati...
PathogenSurveillance: an automated pipeline for population genomic analyses and pathogen identification. #PathogenSurveillance #Nextflow #Bioinformatics #WGS #PathogenIdentification @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 9, 2025 at 5:49 PM
PathogenSurveillance: an automated pipeline for population genomic analyses and pathogen identification. #PathogenSurveillance #Nextflow #Bioinformatics #WGS #PathogenIdentification @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
igv-reports: Embedding interactive genomic visualizations in HTML reports to aid variant review. #IGV #InteractiveVisualizationss #GeneticVariants #VariantReview @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 9, 2025 at 4:00 PM
igv-reports: Embedding interactive genomic visualizations in HTML reports to aid variant review. #IGV #InteractiveVisualizationss #GeneticVariants #VariantReview @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Comparative analysis of single-stranded and non-canonical DNA formation in human and other ape cells with telomere-to-telomere genomes. #NonCanonicalDNA #NonB_DNA #GenomeRegulation @biorxivpreprint.bsky.social 🧪🧬
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 9, 2025 at 10:15 AM
Comparative analysis of single-stranded and non-canonical DNA formation in human and other ape cells with telomere-to-telomere genomes. #NonCanonicalDNA #NonB_DNA #GenomeRegulation @biorxivpreprint.bsky.social 🧪🧬
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 9, 2025 at 9:31 AM
Assembling unmapped reads reveals hidden variation in South Asian genomes. #GenomeVariation #UnmappedReads #SouthAsianPopulation @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 9, 2025 at 8:32 AM
Assembling unmapped reads reveals hidden variation in South Asian genomes. #GenomeVariation #UnmappedReads #SouthAsianPopulation @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Site-specific DNA insertion into the human genome with engineered recombinases. #GenomeEditing #DNAinsertion #LargeSerineRecombinases @natbiotech.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...

November 8, 2025 at 8:05 PM
Site-specific DNA insertion into the human genome with engineered recombinases. #GenomeEditing #DNAinsertion #LargeSerineRecombinases @natbiotech.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Curating 16S rRNA databases enhances taxonomic accuracy and computational efficiency in microbial profiling. #Metabarcoding #16S #rRNA #BacterialGenomes #SequenceDatabases #DatabaseCuration #Genomics #Bioinformatics @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 8, 2025 at 7:05 PM
Curating 16S rRNA databases enhances taxonomic accuracy and computational efficiency in microbial profiling. #Metabarcoding #16S #rRNA #BacterialGenomes #SequenceDatabases #DatabaseCuration #Genomics #Bioinformatics @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Beyond reference bias: Making pangenomes accessible with PangyPlot. #GenomeBrowser #PangenomeGraphs #GeneAnnotations #Visualization #Genomics #Bioinformatics @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
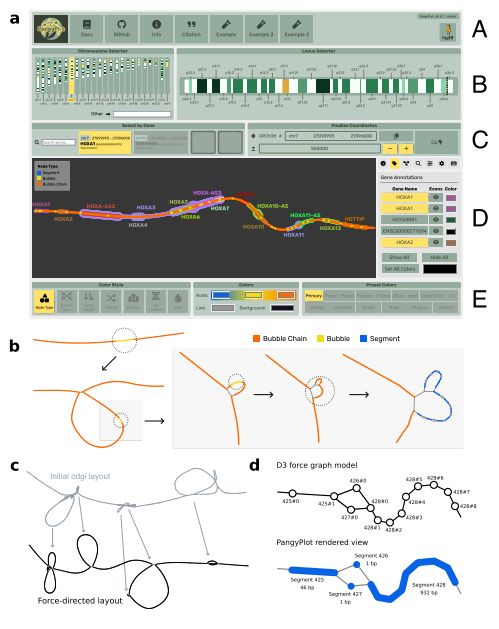
November 8, 2025 at 5:31 PM
Beyond reference bias: Making pangenomes accessible with PangyPlot. #GenomeBrowser #PangenomeGraphs #GeneAnnotations #Visualization #Genomics #Bioinformatics @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
RNA stability enhancers for durable base-modified mRNA therapeutics. #RNAstability #mRNAtherapeutics @natbiotech.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...

November 8, 2025 at 4:01 PM
RNA stability enhancers for durable base-modified mRNA therapeutics. #RNAstability #mRNAtherapeutics @natbiotech.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
A comprehensive view of somatic mosaicism by single-cell DNA analysis. #SingleCell #scDNAseq #SomaticMosaicism @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 8, 2025 at 3:25 PM
A comprehensive view of somatic mosaicism by single-cell DNA analysis. #SingleCell #scDNAseq #SomaticMosaicism @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Defining the Mycobacterium tuberculosis Pangenome and Suggestions for a New Composite Reference Sequence. #MycobacteriumTuberculosis #Pangenome #ReferenceSequence @biorxivpreprint.bsky.social 🧪
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 8, 2025 at 11:49 AM
Defining the Mycobacterium tuberculosis Pangenome and Suggestions for a New Composite Reference Sequence. #MycobacteriumTuberculosis #Pangenome #ReferenceSequence @biorxivpreprint.bsky.social 🧪
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Generating long deletions across the genome with pooled paired prime editing screens. #GenomeDeletions #LongDeletions #PairedPrimeEditing #Genomics @biorxivpreprint.bsky.social 🧪🧬
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 8, 2025 at 10:15 AM
Generating long deletions across the genome with pooled paired prime editing screens. #GenomeDeletions #LongDeletions #PairedPrimeEditing #Genomics @biorxivpreprint.bsky.social 🧪🧬
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Svirlpool: structural variant detection from long read sequencing by local assembly. #LongRead #Sequencing #LRS #StructuralVariants #SVs @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 8, 2025 at 9:31 AM
Svirlpool: structural variant detection from long read sequencing by local assembly. #LongRead #Sequencing #LRS #StructuralVariants #SVs @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
A multi-tissue atlas of genetic regulatory effects in sheep. #SheepGenome #GeneticRegulation #GeneticVariants #Genomics #FarmGTEX #SheepGTEX @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 8, 2025 at 8:32 AM
A multi-tissue atlas of genetic regulatory effects in sheep. #SheepGenome #GeneticRegulation #GeneticVariants #Genomics #FarmGTEX #SheepGTEX @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
PanSpace: Fast and Scalable Indexing for Massive Bacterial Databases. #BacterialDatabase #BacterialGenomes #SequeceIndexing #Kmers #Genomics #Bioinformatics @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 7, 2025 at 8:05 PM
PanSpace: Fast and Scalable Indexing for Massive Bacterial Databases. #BacterialDatabase #BacterialGenomes #SequeceIndexing #Kmers #Genomics #Bioinformatics @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
A phylogenetic estimate of canine retrotransposition rates based on genome assembly comparisons. #CanineGenomes #Retrotransposon #LINE1 #SINE #GenomeAssembly #GenomeEvolution #Genomics @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 7, 2025 at 7:05 PM
A phylogenetic estimate of canine retrotransposition rates based on genome assembly comparisons. #CanineGenomes #Retrotransposon #LINE1 #SINE #GenomeAssembly #GenomeEvolution #Genomics @biorxivpreprint.bsky.social 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Establishing benchmarks for quantitative mapping of m6A by Nanopore Direct RNA Sequencing. #DirectRNAseq #RNAsequencing #RNAmodifications #m6A #Genomics #Bioinformatics @nanoporetech.com 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 7, 2025 at 6:05 PM
Establishing benchmarks for quantitative mapping of m6A by Nanopore Direct RNA Sequencing. #DirectRNAseq #RNAsequencing #RNAmodifications #m6A #Genomics #Bioinformatics @nanoporetech.com 🧪🧬 🖥️
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Unraveling yeast diversity in food fermentation using ITS1-2 amplicon-based metabarcoding. #Metabarcoding #Metagenomics #Yeast #FoodFermentation @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 7, 2025 at 5:32 PM
Unraveling yeast diversity in food fermentation using ITS1-2 amplicon-based metabarcoding. #Metabarcoding #Metagenomics #Yeast #FoodFermentation @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
mRNA 3′ UTRs direct microRNA degradation to participate in imprinted gene networks and regulate growth. #miRNA #miRNAdegratdation #3UTR @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
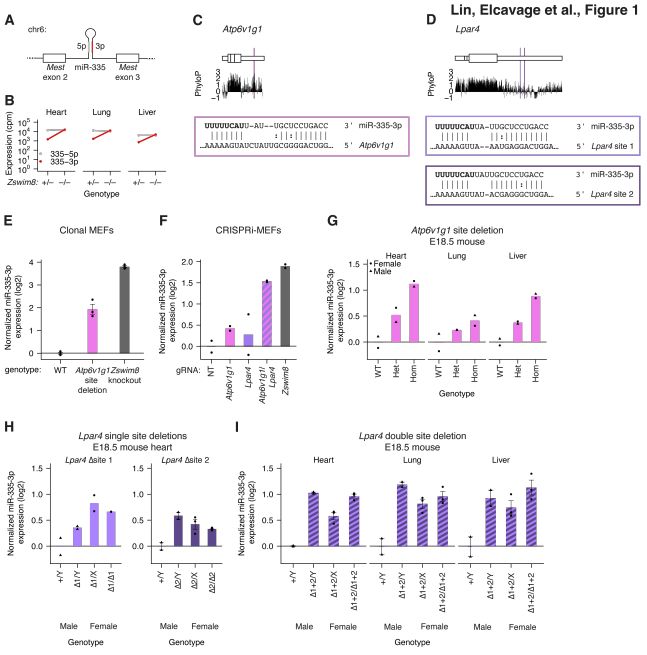
November 7, 2025 at 2:49 PM
mRNA 3′ UTRs direct microRNA degradation to participate in imprinted gene networks and regulate growth. #miRNA #miRNAdegratdation #3UTR @biorxivpreprint.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Establishing benchmarks for quantitative mapping of m6A by Nanopore Direct RNA Sequencing. #DirectRNAseq #Sequencing #RNAmodifications @nanoporetech.com
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 6, 2025 at 11:30 AM
Establishing benchmarks for quantitative mapping of m6A by Nanopore Direct RNA Sequencing. #DirectRNAseq #Sequencing #RNAmodifications @nanoporetech.com
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
RNAcentral in 2026: Genes and literature integration. #nonCodingRNA #ncRNA #Genomics @biorxiv-bioinfo.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 6, 2025 at 10:15 AM
RNAcentral in 2026: Genes and literature integration. #nonCodingRNA #ncRNA #Genomics @biorxiv-bioinfo.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
BINSEQ: A Family of High-Performance Binary Formats for Nucleotide Sequences. #DataFormats #Sequencing #Genomics #Bioinformatics @biorxiv-bioinfo.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

November 6, 2025 at 9:31 AM
BINSEQ: A Family of High-Performance Binary Formats for Nucleotide Sequences. #DataFormats #Sequencing #Genomics #Bioinformatics @biorxiv-bioinfo.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

