https://www.mpg.de/lise-meitner-excellence-program
Programme by the @maxplanckgesellschaft designed to attract and specifically promote exceptionally qualified female #scientists
deadline April 08th, 2025.
note that you should contact […]
https://www.mpg.de/lise-meitner-excellence-program
Programme by the @maxplanckgesellschaft designed to attract and specifically promote exceptionally qualified female #scientists
deadline April 08th, 2025.
note that you should contact […]
It shows: People still #trust scientists and support an active role of scientists in society and policy-making. #OpenAccess available here: www.nature.com/articles/s41... @natureportfolio.bsky.social
(1/13)

It shows: People still #trust scientists and support an active role of scientists in society and policy-making. #OpenAccess available here: www.nature.com/articles/s41... @natureportfolio.bsky.social
(1/13)
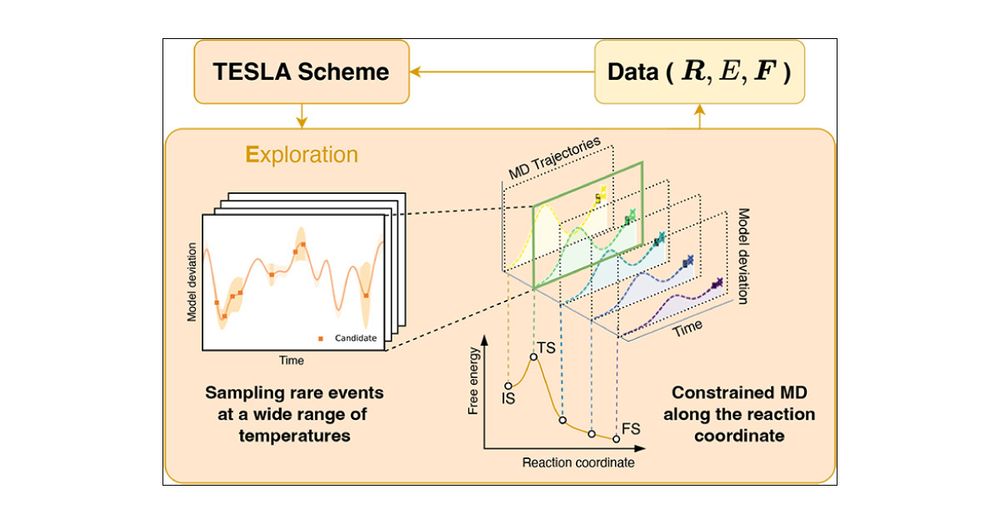

#compchem
www.nature.com/articles/s43...

#compchem
www.nature.com/articles/s43...


in our group in the field of computational and theoretical chemistry
Feel free to forward the info and please apply via
phdheidelberg@gmail.com
in our group in the field of computational and theoretical chemistry
Feel free to forward the info and please apply via
phdheidelberg@gmail.com
I created a starter pack for chemical and biomolecular engineers! Let me know if you want to be added to the list #chemE go.bsky.app/DLGAANj
I created a starter pack for chemical and biomolecular engineers! Let me know if you want to be added to the list #chemE go.bsky.app/DLGAANj

go.bsky.app/Bmp4DHn
go.bsky.app/Bmp4DHn
Made a starter pack for computational chemists, let me know who I missed please
@jchodera.bsky.social
@olexandr.bsky.social
@jelfschem.bsky.social
Made a starter pack for computational chemists, let me know who I missed please
@jchodera.bsky.social
@olexandr.bsky.social
@jelfschem.bsky.social

