Microfluidics, synthetic biology, neurodegeneration, tissue engineering.
I want to develop better in vitro disease models.
Currently looking for positions in the biotech sector in Switzerland / France / Belgium.
Also DM, TTRPG enthusiast.
Have the OFF transceivers express P-cad, and the ON transceiver N-cad.
This just requires a NOT gate downstream of synNotch.
...We have the plasmids already cloned, now we just need a brave student or postdoc to use them!

Have the OFF transceivers express P-cad, and the ON transceiver N-cad.
This just requires a NOT gate downstream of synNotch.
...We have the plasmids already cloned, now we just need a brave student or postdoc to use them!
To better understand how our in vitro results compared to in silico ones, we used a visualization approach with a “morphospace” framework, inspired by @ricardsole.bsky.social

To better understand how our in vitro results compared to in silico ones, we used a visualization approach with a “morphospace” framework, inspired by @ricardsole.bsky.social

So, what about enforcing the segregation of the “growing tip” region?
=> We found that inducing N-cad upon transceivers activation (so, in the “structural support” region) would lead to better segregation! (We are not sure why)

So, what about enforcing the segregation of the “growing tip” region?
=> We found that inducing N-cad upon transceivers activation (so, in the “structural support” region) would lead to better segregation! (We are not sure why)
Here, we believe we share the first screen of this type, as we had to look for inspiration in many publications!
Our two best hits targeted actomyosin contractility: constitutively active RHOA & MLCK.

Here, we believe we share the first screen of this type, as we had to look for inspiration in many publications!
Our two best hits targeted actomyosin contractility: constitutively active RHOA & MLCK.
(We do not yet know why.)
p21 was the only effector which both decreased tissue growth and fluidity, so we decided to build upon its induction.

(We do not yet know why.)
p21 was the only effector which both decreased tissue growth and fluidity, so we decided to build upon its induction.
We found some promising ones, like p53.
… but there was a hidden issue with those…

We found some promising ones, like p53.
… but there was a hidden issue with those…

… and this worked great! (...before physics were completely parametrized!)
(color code mistake here, the growing tip is blue instead of gray!)
… and this worked great! (...before physics were completely parametrized!)
(color code mistake here, the growing tip is blue instead of gray!)
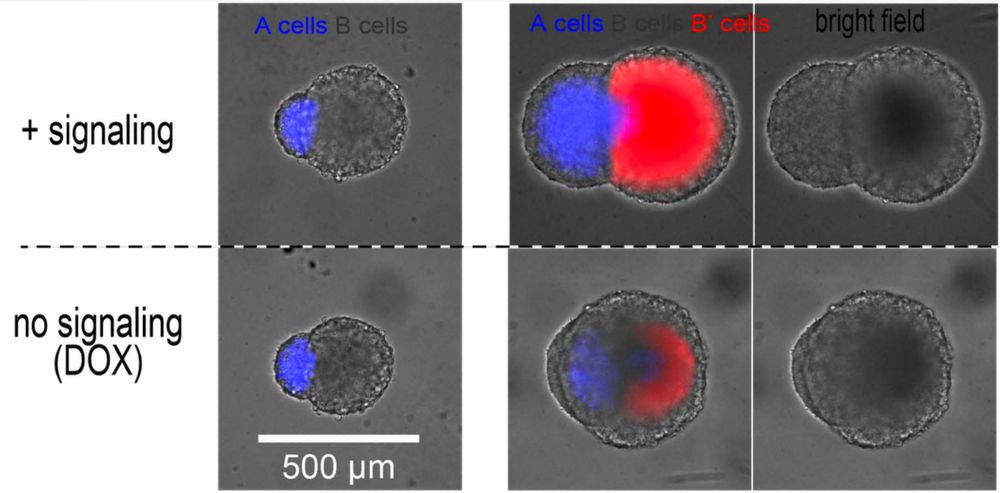
We got the idea that if transceivers changed their properties based on their activation status, this could result in tissue elongation.
For that, the cells must only proliferate when OFF (grey), and become collectively very rigid when ON (red).

We got the idea that if transceivers changed their properties based on their activation status, this could result in tissue elongation.
For that, the cells must only proliferate when OFF (grey), and become collectively very rigid when ON (red).
In this animation, a spheroid of “sender” cells (red+green=yellow) is fused with a spheroid of “transceiver” cells. Activated transceivers become green.
In this animation, a spheroid of “sender” cells (red+green=yellow) is fused with a spheroid of “transceiver” cells. Activated transceivers become green.
We were very lucky… We could just take advantage of another circuit we developed in the lab, the “transceiver” circuit!
(here: www.nature.com/articles/s41467-024-53078-8)

We were very lucky… We could just take advantage of another circuit we developed in the lab, the “transceiver” circuit!
(here: www.nature.com/articles/s41467-024-53078-8)
A combination of local material addition (to the tip) with mechanical resistance to rounding forces (of the bulk).

A combination of local material addition (to the tip) with mechanical resistance to rounding forces (of the bulk).
First, of course, we got our initial inspiration from the literature on axial elongation.
Notably this wonderful 2018 publication from @campaslab.bsky.social where the team demonstrated how tissue fluidity supports axial elongation in the zebrafish embryo!

First, of course, we got our initial inspiration from the literature on axial elongation.
Notably this wonderful 2018 publication from @campaslab.bsky.social where the team demonstrated how tissue fluidity supports axial elongation in the zebrafish embryo!
Here is a 20-ish posts summary, check my repost of Leonardo’s recap for a shorter version 😉

Here is a 20-ish posts summary, check my repost of Leonardo’s recap for a shorter version 😉
biorxiv.org/content/10.1101/2024.12.11.627621v1
We share the first synthetic gene circuit guiding the self-organization of mammalian tissues in elongating structures by dynamically tuning growth, viscosity and adhesion.
biorxiv.org/content/10.1101/2024.12.11.627621v1
We share the first synthetic gene circuit guiding the self-organization of mammalian tissues in elongating structures by dynamically tuning growth, viscosity and adhesion.
=> As distal parts of rod-shaped aggregates are less dense., we get "free" spatial patterning in 3D!
=> Some interesting potential for feedback loops, if this circuit controls tissue physics...

=> As distal parts of rod-shaped aggregates are less dense., we get "free" spatial patterning in 3D!
=> Some interesting potential for feedback loops, if this circuit controls tissue physics...



This is true in 2D and 3D, in mammalian cell fibroblasts and mouse stem cells.


This is true in 2D and 3D, in mammalian cell fibroblasts and mouse stem cells.

