

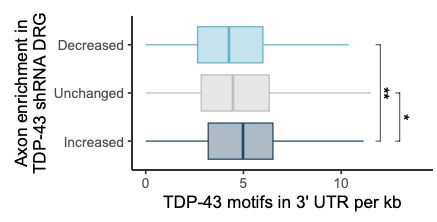










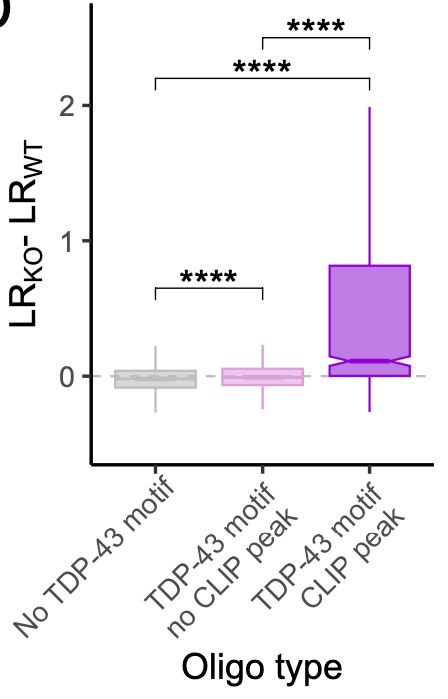



making them TDP-43-sensitive?

making them TDP-43-sensitive?
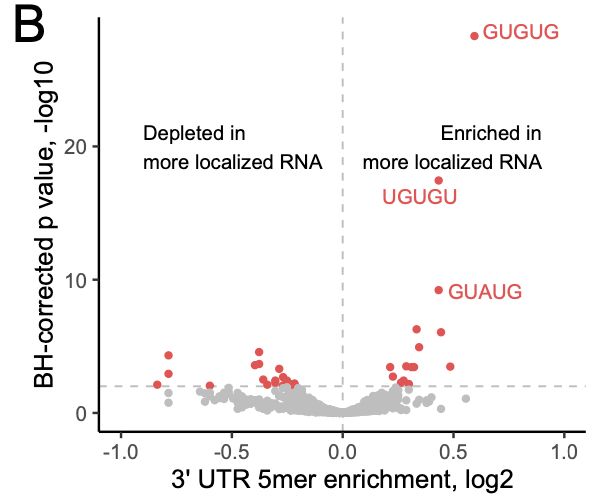







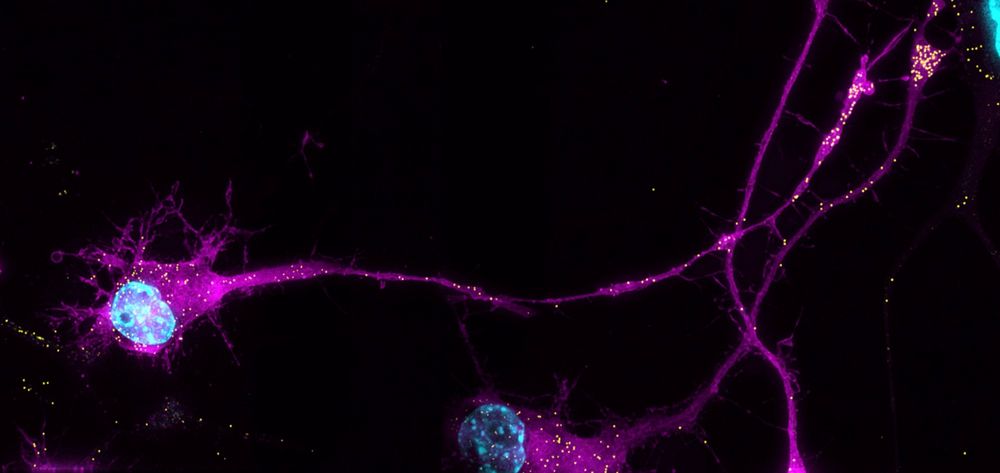

A. extremely arrogant about my own writing
B. an idiot
C. both

A. extremely arrogant about my own writing
B. an idiot
C. both
Apply here: cu.taleo.net/careersectio...

Apply here: cu.taleo.net/careersectio...

[fin] <-- (zebrafish joke)

[fin] <-- (zebrafish joke)



