One of the perks of living in Belgium: epic trails basically in your backyard 🇧🇪🌳
There are a couple of cool two-day hikes here as well which were (rather painful😅) training sessions for our Norway adventure 🇳🇴🏔️
jhsmit.org/hikes/hikema...
#hiking
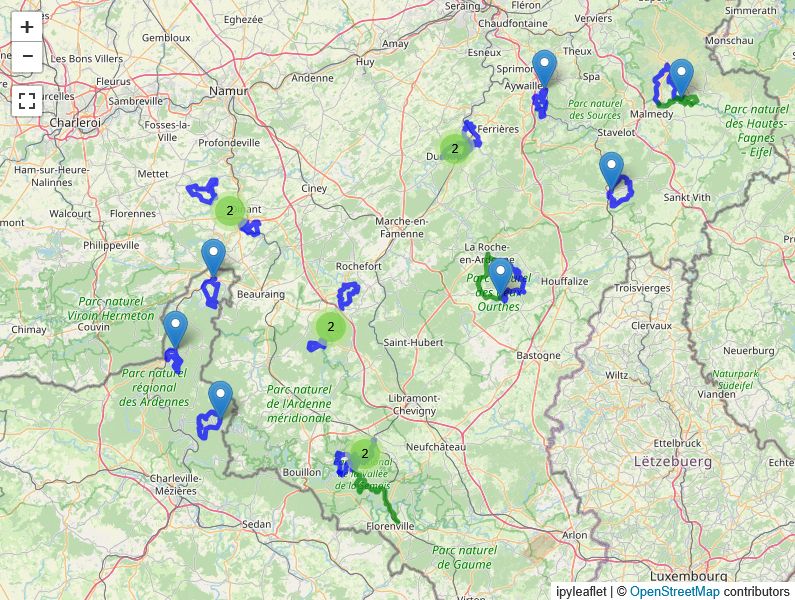
One of the perks of living in Belgium: epic trails basically in your backyard 🇧🇪🌳
There are a couple of cool two-day hikes here as well which were (rather painful😅) training sessions for our Norway adventure 🇳🇴🏔️
jhsmit.org/hikes/hikema...
#hiking

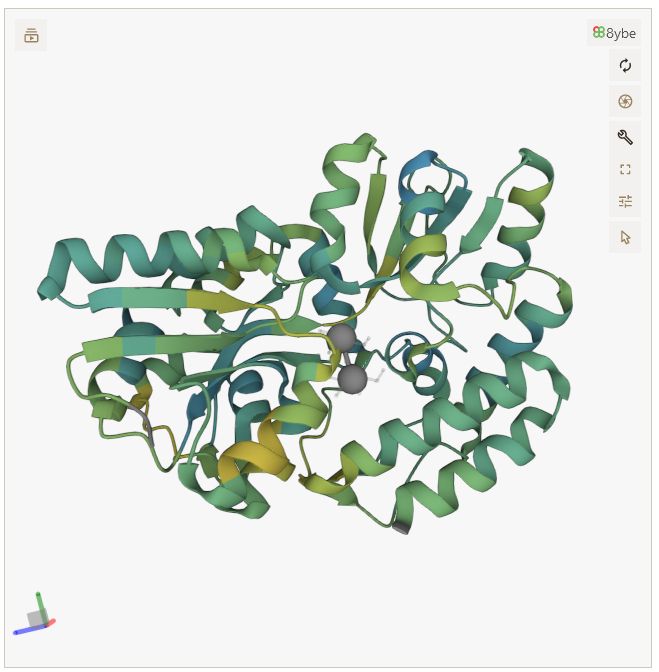
Geometric Algebra feels like magic 🪄; I don't understand it but surely its very powerful 💪
Live example on pycafe: py.cafe/jhsmit/ipymo...
Geometric Algebra feels like magic 🪄; I don't understand it but surely its very powerful 💪
Live example on pycafe: py.cafe/jhsmit/ipymo...
ipymolstar repo: github.com/jhsmit/ipymo...
Molviewspec: molstar.org/mol-view-spec/
ipymolstar repo: github.com/jhsmit/ipymo...
Molviewspec: molstar.org/mol-view-spec/
A cute little boy arrived in our lives early december
Mom & dad are very tired but happy 😃
birth card illustration by Tintenbaum
tintenbaum.com

A cute little boy arrived in our lives early december
Mom & dad are very tired but happy 😃
birth card illustration by Tintenbaum
tintenbaum.com
Upload your .csv with residue/value data, select data column, colormap and norm to interactively color your protein model.
Then, add title/description and click the button to open a shareable view with url-encoded data and settings.
Upload your .csv with residue/value data, select data column, colormap and norm to interactively color your protein model.
Then, add title/description and click the button to open a shareable view with url-encoded data and settings.
Try it on pycafe: py.cafe/jhsmit/ipymo...
Try it on pycafe: py.cafe/jhsmit/ipymo...
huggingface.co/spaces/Jhsmi...
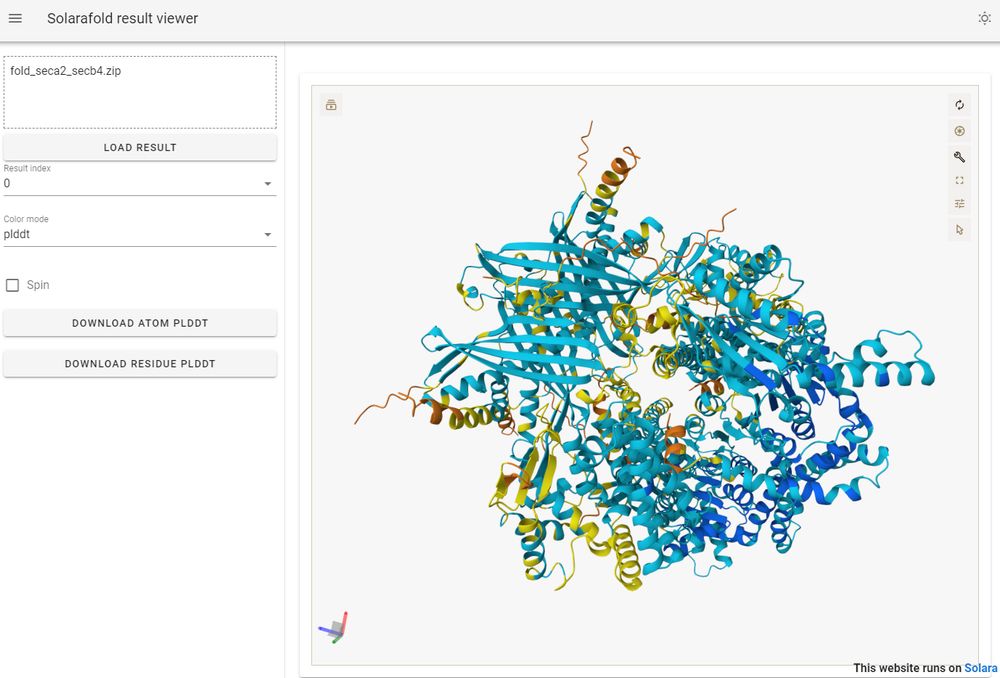
huggingface.co/spaces/Jhsmi...
`pip install "ipymolstar==0.0.4"`
github.com/Jhsmit/ipymo...
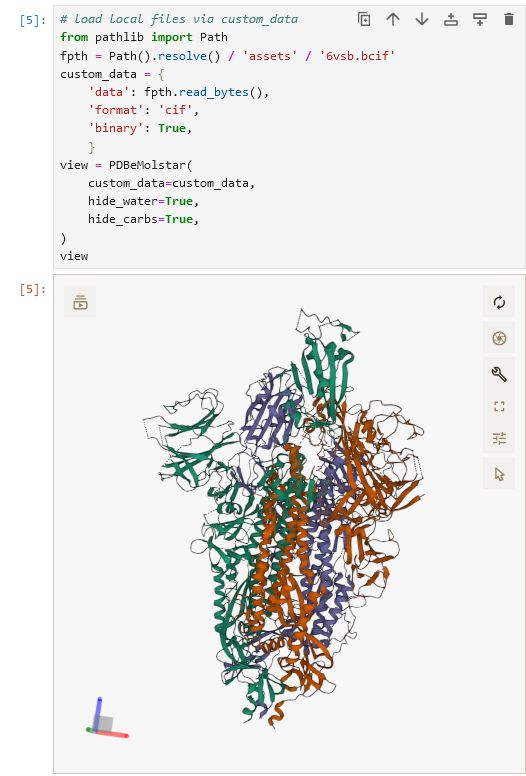
`pip install "ipymolstar==0.0.4"`
github.com/Jhsmit/ipymo...