Jacob Corn
@jcornlab.bsky.social
Genome editing, functional genomics, and cells figuring out how to eat themselves without dying. Professor of Genome Biology at ETH Zürich.
This was a great collaboration with the lab of Virginijus Siksnys!
October 28, 2025 at 1:29 PM
This was a great collaboration with the lab of Virginijus Siksnys!
This was an incredible collaboration with the labs of Randall Platt, Gerald Schwank, Steve Jackson, Helmuth Gehart, and Timm Schroeder. Special thanks also to the NOMIS Foundation who supported some of the very ambitious experiments.
October 28, 2025 at 7:33 AM
This was an incredible collaboration with the labs of Randall Platt, Gerald Schwank, Steve Jackson, Helmuth Gehart, and Timm Schroeder. Special thanks also to the NOMIS Foundation who supported some of the very ambitious experiments.
Definitely true that epigenetic state can affect repair. We try to control for this by editing in highly expressed housekeeping genes with approx equal epigenetic states in all cells. And HDR is just one change. There are stereotyped differences in categorical and molecular end joining outcomes.
October 24, 2025 at 1:46 PM
Definitely true that epigenetic state can affect repair. We try to control for this by editing in highly expressed housekeeping genes with approx equal epigenetic states in all cells. And HDR is just one change. There are stereotyped differences in categorical and molecular end joining outcomes.
Generally true, but I'm pretty sure we're not seeing sampling error. If that were true, then equally populated cell subtypes would have similar differential outcomes. Instead, we can recover cell type solely by clustering on differential outcomes! And "stemmy" cells share outcomes across lineages.
October 24, 2025 at 1:43 PM
Generally true, but I'm pretty sure we're not seeing sampling error. If that were true, then equally populated cell subtypes would have similar differential outcomes. Instead, we can recover cell type solely by clustering on differential outcomes! And "stemmy" cells share outcomes across lineages.
There's some crazy stuff in this preprint. For example, did you know that some neuronal subsets actually do HDR with surprising efficiency?
October 24, 2025 at 10:05 AM
There's some crazy stuff in this preprint. For example, did you know that some neuronal subsets actually do HDR with surprising efficiency?
I’m still learning Bluesky, so now tagging people I mis-tagged. Sorry! @charlesyeh.bsky.social @albertomarin.bsky.social @fedeteloni.bsky.social @gerlichlab.bsky.social
February 12, 2025 at 4:38 PM
I’m still learning Bluesky, so now tagging people I mis-tagged. Sorry! @charlesyeh.bsky.social @albertomarin.bsky.social @fedeteloni.bsky.social @gerlichlab.bsky.social
If you’re interested in how loop-extruding cohesin drives the homology search during HR, check out also this new preprint from albertomarin.bsky.social in the Taekjip Ha lab (doi.org/10.1101/2025...)
doi.org
February 12, 2025 at 4:04 PM
If you’re interested in how loop-extruding cohesin drives the homology search during HR, check out also this new preprint from albertomarin.bsky.social in the Taekjip Ha lab (doi.org/10.1101/2025...)
Curious about how cohesin guides homology search via loops & sister chromatid linkages? Check out this new preprint by fedeteloni.bsky.social & gerlichlab.bsky.social! 👉 tinyurl.com/DSBcohesin
tinyurl.com
February 12, 2025 at 4:04 PM
Curious about how cohesin guides homology search via loops & sister chromatid linkages? Check out this new preprint by fedeteloni.bsky.social & gerlichlab.bsky.social! 👉 tinyurl.com/DSBcohesin
This project was driven by amazing graduate student charlesyeh.bsky.social, with lots of amazing collaborators over the years.
Charles Yeh (@charlesyeh.bsky.social)
charlesyeh.bsky.social
February 12, 2025 at 4:03 PM
This project was driven by amazing graduate student charlesyeh.bsky.social, with lots of amazing collaborators over the years.
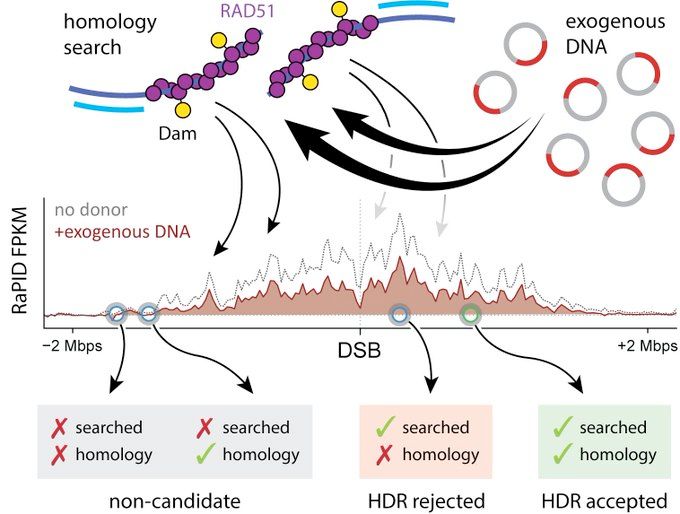
Altogether, proximity is key and search normally stays around the DSB. When it comes choosing a donor, only these sequences are candidates.
February 12, 2025 at 4:03 PM
Altogether, proximity is key and search normally stays around the DSB. When it comes choosing a donor, only these sequences are candidates.
During genome editing, exogenous donor DNAs soak up homology search during HDR. Even if they have no sequence relationship to the cut site at all. They actually steal search from the genome!
February 12, 2025 at 4:02 PM
During genome editing, exogenous donor DNAs soak up homology search during HDR. Even if they have no sequence relationship to the cut site at all. They actually steal search from the genome!
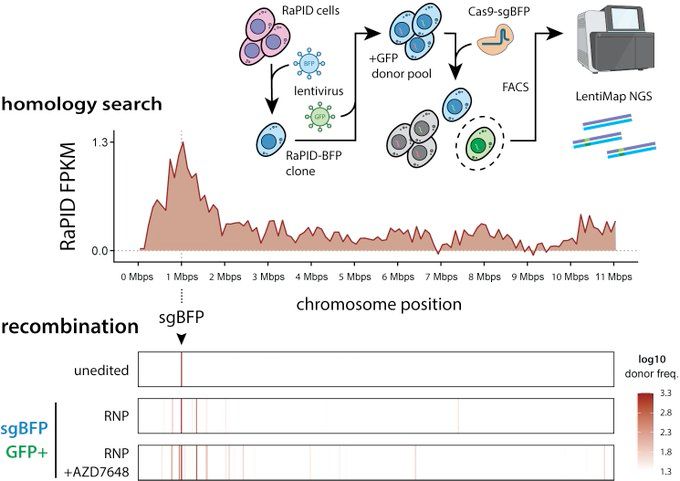
Only 3D-nearby those sequences that are searched (i.e., candidates) are chosen as HDR donors! Donors too far away are never seen and never used.
February 12, 2025 at 4:02 PM
Only 3D-nearby those sequences that are searched (i.e., candidates) are chosen as HDR donors! Donors too far away are never seen and never used.
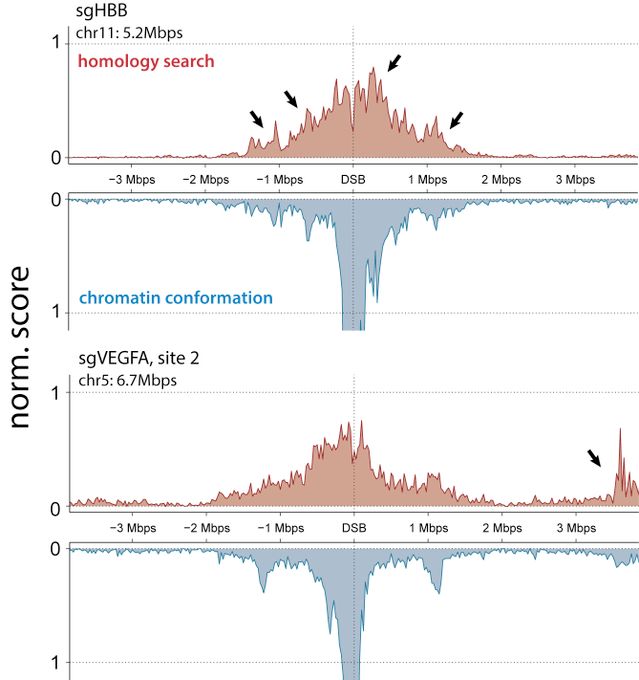
Search is constrained by chromatin conformation. Check out the related preprints from fedeteloni.bsky.social
and albertomarin.bsky.social for a much deeper dive (see later in thread).
and albertomarin.bsky.social for a much deeper dive (see later in thread).
February 12, 2025 at 4:02 PM
Search is constrained by chromatin conformation. Check out the related preprints from fedeteloni.bsky.social
and albertomarin.bsky.social for a much deeper dive (see later in thread).
and albertomarin.bsky.social for a much deeper dive (see later in thread).
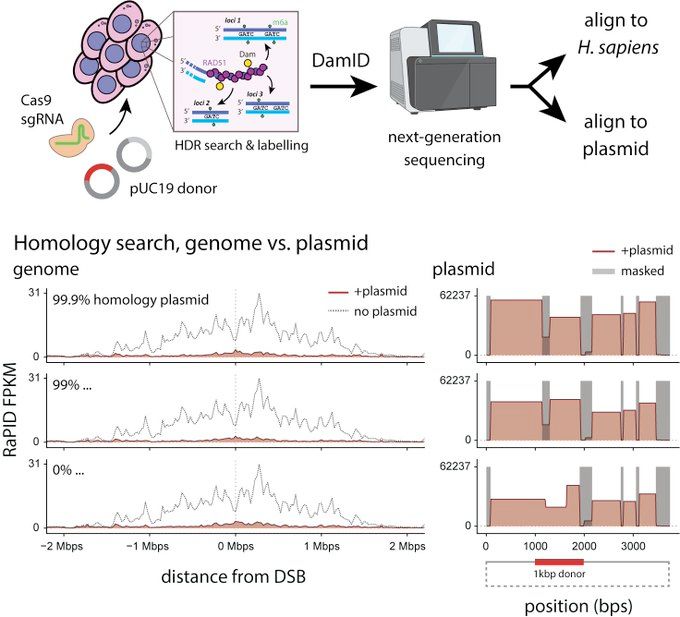
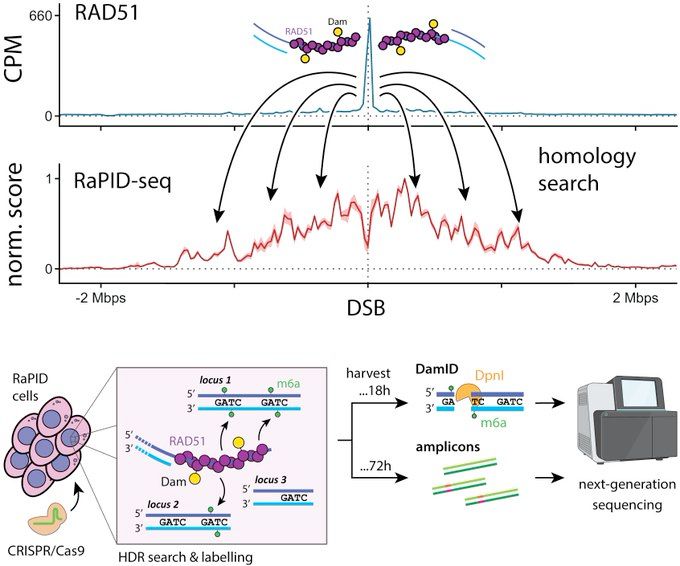
We call this "RaPID-seq". It unveils a new dimension of DNA repair, telling us what is searched during HDR. In other words, what are the candidates?
February 12, 2025 at 4:00 PM
We call this "RaPID-seq". It unveils a new dimension of DNA repair, telling us what is searched during HDR. In other words, what are the candidates?
This project was led by
@matthiasmuhar.bsky.social, @jakobfarnung.bsky.social Martina Cernakova, and Raphael Hofmann.
@matthiasmuhar.bsky.social, @jakobfarnung.bsky.social Martina Cernakova, and Raphael Hofmann.
January 30, 2025 at 12:35 PM
This project was led by
@matthiasmuhar.bsky.social, @jakobfarnung.bsky.social Martina Cernakova, and Raphael Hofmann.
@matthiasmuhar.bsky.social, @jakobfarnung.bsky.social Martina Cernakova, and Raphael Hofmann.
Thanks to amazing collaborators in the Schulman, Jinek, and Jessberger labs. And most of all to Jeff Bode's lab, who has been with this every step of the way. @micharapelab.bsky.social wrote a News and Views that explains our work far better than I ever could. www.nature.com/articles/d41...

Signs of damage that drive protein degradation
How the stress-response machinery eliminates damaged proteins.
www.nature.com
January 30, 2025 at 12:33 PM
Thanks to amazing collaborators in the Schulman, Jinek, and Jessberger labs. And most of all to Jeff Bode's lab, who has been with this every step of the way. @micharapelab.bsky.social wrote a News and Views that explains our work far better than I ever could. www.nature.com/articles/d41...

