Jacob Corn
@jcornlab.bsky.social
Genome editing, functional genomics, and cells figuring out how to eat themselves without dying. Professor of Genome Biology at ETH Zürich.
Congrats to Moritz Schlapansky, who successfully defended his PhD thesis on Friday! You can read his work on "scOUT-seq" profiling of single cell transcriptomes + editing outcomes in millions of cells and living mice at www.biorxiv.org/content/10.1...

November 10, 2025 at 7:56 AM
Congrats to Moritz Schlapansky, who successfully defended his PhD thesis on Friday! You can read his work on "scOUT-seq" profiling of single cell transcriptomes + editing outcomes in millions of cells and living mice at www.biorxiv.org/content/10.1...
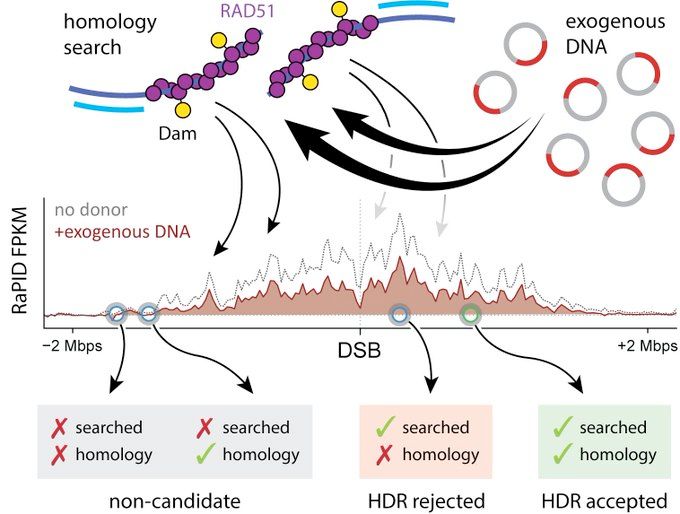
Altogether, proximity is key and search normally stays around the DSB. When it comes choosing a donor, only these sequences are candidates.
February 12, 2025 at 4:03 PM
Altogether, proximity is key and search normally stays around the DSB. When it comes choosing a donor, only these sequences are candidates.
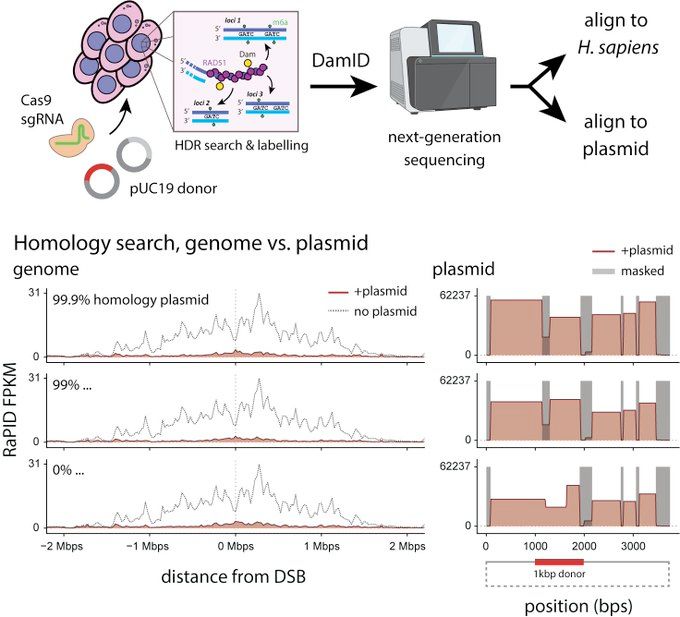
During genome editing, exogenous donor DNAs soak up homology search during HDR. Even if they have no sequence relationship to the cut site at all. They actually steal search from the genome!
February 12, 2025 at 4:02 PM
During genome editing, exogenous donor DNAs soak up homology search during HDR. Even if they have no sequence relationship to the cut site at all. They actually steal search from the genome!
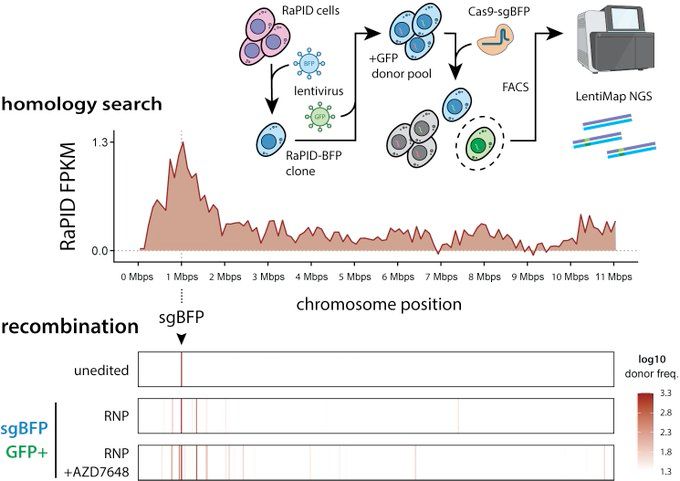
Only 3D-nearby those sequences that are searched (i.e., candidates) are chosen as HDR donors! Donors too far away are never seen and never used.
February 12, 2025 at 4:02 PM
Only 3D-nearby those sequences that are searched (i.e., candidates) are chosen as HDR donors! Donors too far away are never seen and never used.
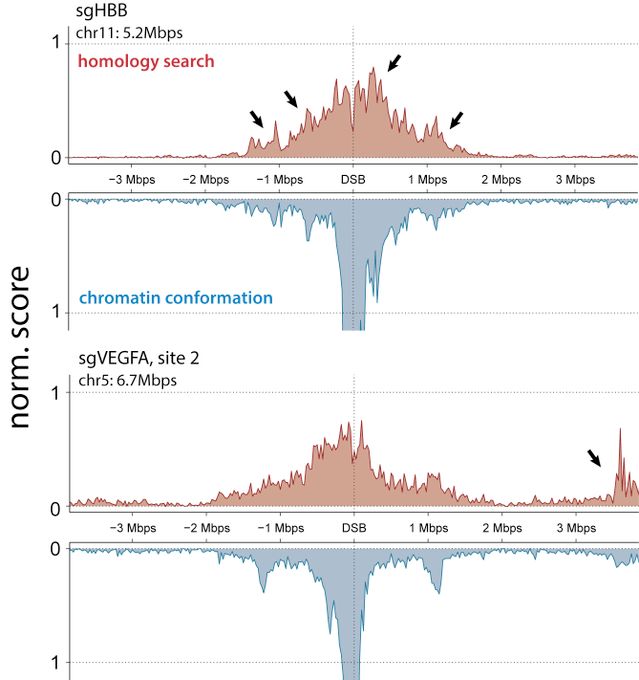
Search is constrained by chromatin conformation. Check out the related preprints from fedeteloni.bsky.social
and albertomarin.bsky.social for a much deeper dive (see later in thread).
and albertomarin.bsky.social for a much deeper dive (see later in thread).
February 12, 2025 at 4:02 PM
Search is constrained by chromatin conformation. Check out the related preprints from fedeteloni.bsky.social
and albertomarin.bsky.social for a much deeper dive (see later in thread).
and albertomarin.bsky.social for a much deeper dive (see later in thread).
We call this "RaPID-seq". It unveils a new dimension of DNA repair, telling us what is searched during HDR. In other words, what are the candidates?
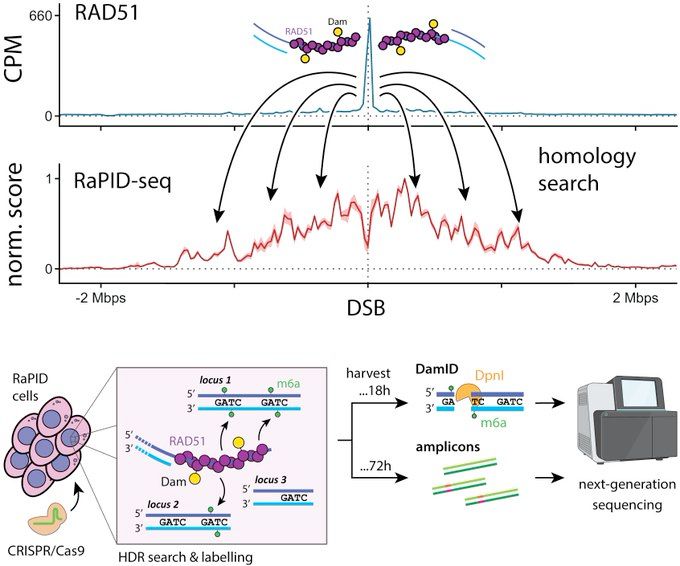
February 12, 2025 at 4:00 PM
We call this "RaPID-seq". It unveils a new dimension of DNA repair, telling us what is searched during HDR. In other words, what are the candidates?

