John Chodera
@jchodera.bsky.social
Achira | http://achira.ai
Research laboratory | http://choderalab.org
Antiviral drug discovery for pandemics | http://asapdiscovery.org
OpenADMET | http://openadmet.org
Employer-mandated disclaimer: http://choderalab.org/disclaimer
Pronouns: he/him
Research laboratory | http://choderalab.org
Antiviral drug discovery for pandemics | http://asapdiscovery.org
OpenADMET | http://openadmet.org
Employer-mandated disclaimer: http://choderalab.org/disclaimer
Pronouns: he/him
Chat, is this good?

October 11, 2025 at 1:53 AM
Chat, is this good?
Excited to be joining the AI for Science Symposium in San Francisco on May 16 organized by ML Foundry!
Check it out here:
mlfoundry.com/ai-for-scien...
Check it out here:
mlfoundry.com/ai-for-scien...

May 2, 2025 at 4:54 PM
Excited to be joining the AI for Science Symposium in San Francisco on May 16 organized by ML Foundry!
Check it out here:
mlfoundry.com/ai-for-scien...
Check it out here:
mlfoundry.com/ai-for-scien...
I have so many of these!

February 21, 2025 at 9:31 PM
I have so many of these!
Oh, have we got coffee cup ideas for you...

February 21, 2025 at 5:11 PM
Oh, have we got coffee cup ideas for you...
As a peek toward where we're headed:
Right now, CADD scientists are forced to use the same model week after week, even if new experimental data says the model is inaccurate.
If we can fine- models, we can exploit that data to systematically improve our predictions week by week!
Right now, CADD scientists are forced to use the same model week after week, even if new experimental data says the model is inaccurate.
If we can fine- models, we can exploit that data to systematically improve our predictions week by week!

February 19, 2025 at 7:36 PM
As a peek toward where we're headed:
Right now, CADD scientists are forced to use the same model week after week, even if new experimental data says the model is inaccurate.
If we can fine- models, we can exploit that data to systematically improve our predictions week by week!
Right now, CADD scientists are forced to use the same model week after week, even if new experimental data says the model is inaccurate.
If we can fine- models, we can exploit that data to systematically improve our predictions week by week!
While this strategy is still highly limited by the constraints of legacy molecular mechanics force fields which were developed for late-1970s era hardware.
Since then, compute per dollar has increased by 160 billion times. We need a totally new approach. Stay tuned for something exciting...
Since then, compute per dollar has increased by 160 billion times. We need a totally new approach. Stay tuned for something exciting...

February 19, 2025 at 7:30 PM
While this strategy is still highly limited by the constraints of legacy molecular mechanics force fields which were developed for late-1970s era hardware.
Since then, compute per dollar has increased by 160 billion times. We need a totally new approach. Stay tuned for something exciting...
Since then, compute per dollar has increased by 160 billion times. We need a totally new approach. Stay tuned for something exciting...
This simple regularization strategy was surprisingly effective at ensuring the reweighted free energy estimates at new parameters matched free energies from expensive new simulations at the new parameter set.
![Figure 5. Fine-tuned Optimized/Reweighted hydration free energies are highly consistent with Optimized/Recalculated hydration free energies and demonstrate statistically significant accuracy improvements over the foundation model espaloma-0.3.2. (A) Correlation plot between Optimized/Reweighted free energies and Optimized/Resimulated free energies show RMSEs of 0.06 kcal mol−1, demonstrate agreement between Zwanzig reweighting used in fine-tuning optimization and the Bennet Acceptance Ratio (BAR) [7], which was used in the free energy recalculation experiments at optimized molecular partial charges. (B) Experimental vs calculated hydration free energy plots by dataset show consistent improvements in the Optimized/Resimulated calculations over the baseline foundation model espaloma0.3.2. (C) Absolute hydration free energy residual CDFs for each data split and experiment show high agreement between residuals of the Optimized/Resimulated (green) and Optimized/Reweighted (orange) data (consistent with panel A) and reliable improvement over the baseline foundation model (green) in all data splits.
From https://doi.org/10.1101/2025.01.06.631610](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:2rajbtnvpcqnm7tzayvgxo5d/bafkreiepu7v4wrcoutdcmh47capp6dfrl2sqcyjwvaoris3q7d6cw3lezu@jpeg)
February 19, 2025 at 7:30 PM
This simple regularization strategy was surprisingly effective at ensuring the reweighted free energy estimates at new parameters matched free energies from expensive new simulations at the new parameter set.
Dominic showed that a regularized low-rank adjustment to the espaloma-0.3 MM foundation simulation model dramatically reduced errors in predicted hydration free energies in the held-out test set, shifting the error CDF far to the left, improving the RMSE by 0.82 kcal/mol--in just ten minutes!

February 19, 2025 at 7:30 PM
Dominic showed that a regularized low-rank adjustment to the espaloma-0.3 MM foundation simulation model dramatically reduced errors in predicted hydration free energies in the held-out test set, shifting the error CDF far to the left, improving the RMSE by 0.82 kcal/mol--in just ten minutes!
But this can be avoided! By adding a regularization term that penalizes effective sample size collapse, optimization takes a different path that maintains the ability of reweighting strategies to estimate accurate free energies for the new model parameters.

February 19, 2025 at 7:30 PM
But this can be avoided! By adding a regularization term that penalizes effective sample size collapse, optimization takes a different path that maintains the ability of reweighting strategies to estimate accurate free energies for the new model parameters.
Dominic also wanted it to be FAST. Could we do this in one step, avoiding need for additional simulations?
Unfortunately, once you start training the LoRA model, the conformational probability density can change rapidly, quickly escaping the region where free energy reweighting gives low errors.
Unfortunately, once you start training the LoRA model, the conformational probability density can change rapidly, quickly escaping the region where free energy reweighting gives low errors.

February 19, 2025 at 7:30 PM
Dominic also wanted it to be FAST. Could we do this in one step, avoiding need for additional simulations?
Unfortunately, once you start training the LoRA model, the conformational probability density can change rapidly, quickly escaping the region where free energy reweighting gives low errors.
Unfortunately, once you start training the LoRA model, the conformational probability density can change rapidly, quickly escaping the region where free energy reweighting gives low errors.
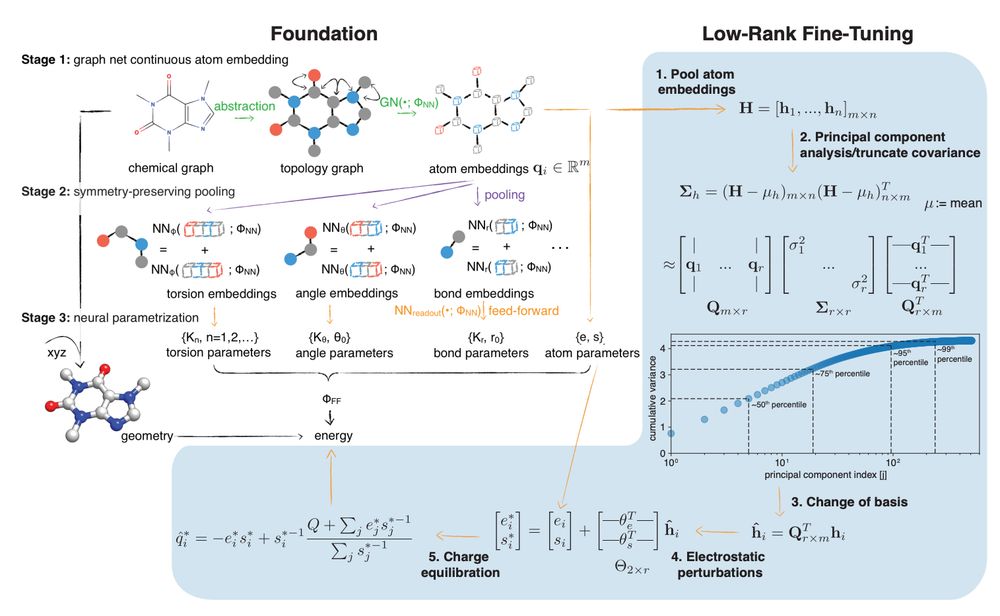
Borrowing ideas from fine-tuning LLMs, Dominic asked whether a low-rank adaptation (LoRA) could map from atom embedding vectors to learned perturbations to the charge-equilibration model parameters (electronegativity and hardness) to modify the small molecule partial charges.
February 19, 2025 at 7:30 PM
Borrowing ideas from fine-tuning LLMs, Dominic asked whether a low-rank adaptation (LoRA) could map from atom embedding vectors to learned perturbations to the charge-equilibration model parameters (electronegativity and hardness) to modify the small molecule partial charges.
For example, hydration free energies can be computed with high precision using alchemical free energy methods with Ken's espaloma-0.3 foundation model, but are relatively inaccurate, with an RMSE ~ 1.72 kcal/mol and a disappointing error cumulative distribution function (CDF).

February 19, 2025 at 7:30 PM
For example, hydration free energies can be computed with high precision using alchemical free energy methods with Ken's espaloma-0.3 foundation model, but are relatively inaccurate, with an RMSE ~ 1.72 kcal/mol and a disappointing error cumulative distribution function (CDF).
What can you do with a foundation simulation model? It's good at many tasks, but may not achieve the performance you want for every application.
Former PhD student Dominic Rufa wondered whether we could fine-tune these models to deliver better performance.
www.linkedin.com/in/dominic-r...
Former PhD student Dominic Rufa wondered whether we could fine-tune these models to deliver better performance.
www.linkedin.com/in/dominic-r...

February 19, 2025 at 7:30 PM
What can you do with a foundation simulation model? It's good at many tasks, but may not achieve the performance you want for every application.
Former PhD student Dominic Rufa wondered whether we could fine-tune these models to deliver better performance.
www.linkedin.com/in/dominic-r...
Former PhD student Dominic Rufa wondered whether we could fine-tune these models to deliver better performance.
www.linkedin.com/in/dominic-r...
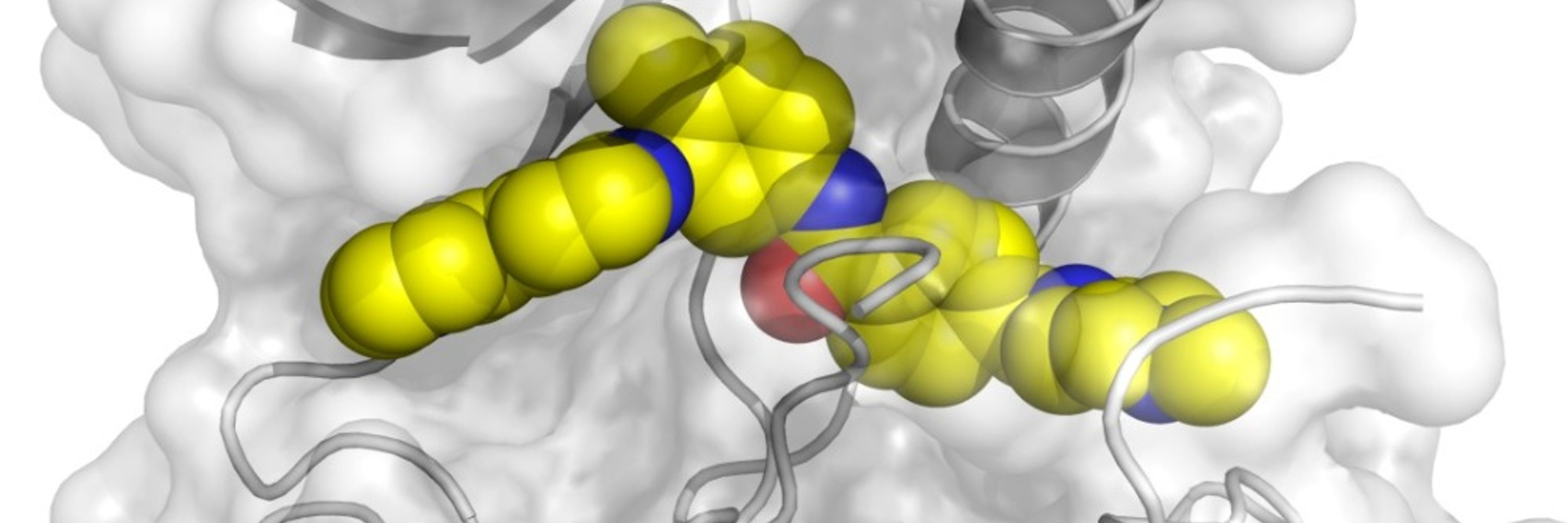
In a tour de force, visiting scientist Ken Takaba [https://kntkb.github.io/] built the first true molecular mechanics foundation simulation model for proteins, nucleic acids, and small molecules that excels at at protein:ligand binding free energy benchmarks.
pubs.rsc.org/en/content/a...
pubs.rsc.org/en/content/a...
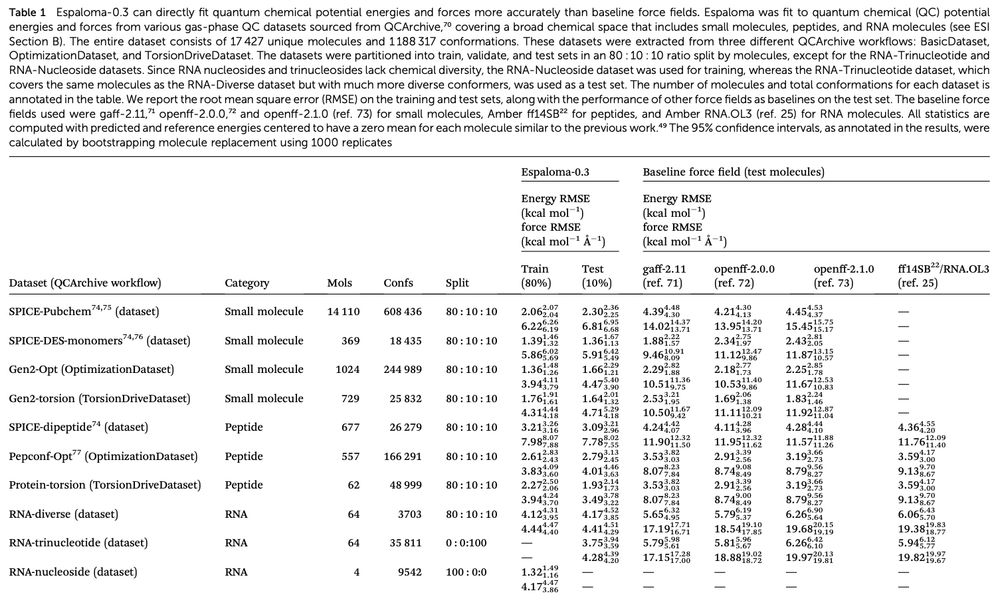

![Fig. 5 espaloma-0.3 can be used for accurate protein–ligand alchemical free energy calculations. (a) Protein–ligand (PL) alchemical free energy calculations were calculated for Tyk2 (10 ns/replica), Cdk2 (10 ns/replica), Mcl1 (15 ns/replica), P38 (20 ns/replica) using a curated PL-benchmark dataset (see ESI Section F†) which comprises 76 ligands in total. The PL structures used to setup the alchemical free energy calculations for each target system is shown. Here, we used Perses 0.10.1 relative free energy calculation infrastructure,110 based on OpenMM 8.0.0,111 to assess the accuracy of espaloma-0.3 and openff-2.1.0 (ref. 73) combined with Amber ff14SB force field22 for comparison. (b) Schematic illustration of the alchemical ligand transformation network for Tyk2. The methyl R-group in the center is alchemically transformed into various R-groups. The binding free energy for each R-group is annotated alongside the respective R-groups. (c) The openff-2.1.0 (ref. 73) with protein parametrized with Amber ff14SB force field (ff14SB + openff-2.1.0) achieves an absolute free energy (DG) RMSE of 1.01 [95% CI: 0.73, 1.33] kcal mol−1. The espaloma-0.3 for predicting valence parameters and partial charges of small molecules combined with Amber ff14SB force field for proteins (ff14SB + espaloma-0.3) achieves an absolute free energy (DG) RMSE of 1.13 [95% CI: 0.86, 1.47] kcal mol−1. Parametrizing small molecule and protein self-consistently with espaloma-0.3 (espaloma-0.3) achieves absolute free energy (DG) RMSE of 1.02 [95% CI: 0.74, 1.37] kcal mol−1 which is comparable to those obtained by (ff14SB + openff-2.1.0) and (ff14SB + espaloma-0.3). All systems were solvated with TIP3P water26 and neutralized with 300 mM NaCl salt using Joung and Cheatham monovalent counterions.29 The light and dark gray regions depict the confidence bounds of 0.5 kcal mol−1 and 1.0 kcal mol−1, respectively.
From https://arxiv.org/abs/2307.07085](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:2rajbtnvpcqnm7tzayvgxo5d/bafkreidltrg2rqvinyk7hkmoex4cxwjukjc2cjaux63p42i5hpqk4jt4om@jpeg)
February 19, 2025 at 7:30 PM
In a tour de force, visiting scientist Ken Takaba [https://kntkb.github.io/] built the first true molecular mechanics foundation simulation model for proteins, nucleic acids, and small molecules that excels at at protein:ligand binding free energy benchmarks.
pubs.rsc.org/en/content/a...
pubs.rsc.org/en/content/a...
Aside: Yuanqing Wang is tremendously talented, and is on the faculty job market right now!
Check out his website: www.wangyq.net
and papers: scholar.google.com/citations?us...
Check out his website: www.wangyq.net
and papers: scholar.google.com/citations?us...

February 19, 2025 at 7:30 PM
Aside: Yuanqing Wang is tremendously talented, and is on the faculty job market right now!
Check out his website: www.wangyq.net
and papers: scholar.google.com/citations?us...
Check out his website: www.wangyq.net
and papers: scholar.google.com/citations?us...
Recently, Yuanqing Wang [https://www.wangyq.net/] (now a Simons and Schmidt Fellow at NYU) demonstrated how molecular mechanics force field construction can be cast as an end-to-end differentiable machine learning problem, presenting espaloma: espaloma.wangyq.net
![Figure 1. Espaloma is an end-to-end differentiable molecular mechanics parameter assignment scheme for arbitrary organic molecules. espaloma (extendable surrogate potential optimized by message-passing) is a modular approach
for directly computing molecular mechanics force field parameters ΦFF from a chemical graph such as a small molecule
or biopolymer via a process that is fully differentiable in the model parameters ΦNN. In Stage 1, a graph neural network is
used to generate continuous latent atom embeddings describing local chemical environments from the chemical graph.
In Stage 2, these atom embeddings are transformed into feature vectors that preserve appropriate symmetries for atom,
bond, angle, and proper/improper torsion inference via Janossy pooling. In Stage 3, molecular mechanics parameters
are directly predicted from these feature vectors using feed-forward neural nets. This parameter assignment process
is performed once per molecular species, allowing the potential energy to be rapidly computed using standard molecular mechanics or molecular dynamics frameworks thereafter. The collection of parameters ΦNN describing the espaloma
model can be considered as the equivalent complete specification of a traditional molecular mechanics force field such as
GAFF [26, 27]/AM1-BCC [28, 29] in that it encodes the equivalent of traditional typing rules, parameter assignment tables,
and even partial charge models. This final stage is modular, and can be easily extended to incorporate additional molecular mechanics parameter classes, such as parameters for a charge-equilibration model (Section 4), point polarizabilities,
or valence-coupling terms for Class II molecular mechanics force fields [30, 31].
Figure from Wang Y, Fass J, and Chodera JD “End-to-End Differentiable Construction of Molecular Mechanics Force Fields. https://arxiv.org/abs/2010.01196](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:2rajbtnvpcqnm7tzayvgxo5d/bafkreifevfhrqqqpyr7aspqxhvjaevijynvuqeopjfbo655ss6ehvxh3y4@jpeg)
February 19, 2025 at 7:30 PM
Recently, Yuanqing Wang [https://www.wangyq.net/] (now a Simons and Schmidt Fellow at NYU) demonstrated how molecular mechanics force field construction can be cast as an end-to-end differentiable machine learning problem, presenting espaloma: espaloma.wangyq.net
Everything is chaos, but I wanted to share some awesome recent science from the lab that hints at where the future of biomolecular simulation is headed:
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.

February 19, 2025 at 7:30 PM
Everything is chaos, but I wanted to share some awesome recent science from the lab that hints at where the future of biomolecular simulation is headed:
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
Foundation simulation models that can be fine-tuned to experimental free energy data to produce systematically more accurate predictions.
In case you want to do something about it:
resist.bot/petitions/PM...
resist.bot/petitions/PM...

January 28, 2025 at 1:20 PM
In case you want to do something about it:
resist.bot/petitions/PM...
resist.bot/petitions/PM...
Happy Caturday to all who celebrate.

December 22, 2024 at 4:57 AM
Happy Caturday to all who celebrate.
Daniel Probst (@skepteis.bsky.social) is up next with "Less is More" at the ML4Molecules Workshop in Berlin, taking his own advice by presenting without the need for corporeal form.

December 6, 2024 at 12:45 PM
Daniel Probst (@skepteis.bsky.social) is up next with "Less is More" at the ML4Molecules Workshop in Berlin, taking his own advice by presenting without the need for corporeal form.
Cecilia Clementi (@cecclementi.bsky.social) kicks off the afternoon session of the ELLIS ML4Molecules Workshop in Berlin!

December 6, 2024 at 12:03 PM
Cecilia Clementi (@cecclementi.bsky.social) kicks off the afternoon session of the ELLIS ML4Molecules Workshop in Berlin!
Excited to speak at the ELLIS ML4Molecules Workshop 2024 in Berlin!
moleculediscovery.github.io/workshop2024/
moleculediscovery.github.io/workshop2024/

December 6, 2024 at 8:08 AM
Excited to speak at the ELLIS ML4Molecules Workshop 2024 in Berlin!
moleculediscovery.github.io/workshop2024/
moleculediscovery.github.io/workshop2024/




