Bloom lab
@jbloomlab.bsky.social
Lab studying molecular evolution of proteins and viruses. Affiliated with Fred Hutch & HHMI.
Opinions are my own and do not reflect those of my employer.
https://jbloomlab.org/
Opinions are my own and do not reflect those of my employer.
https://jbloomlab.org/
Above visualizations just scratch surface of data: there is tremendous heterogeneity across sera from different individuals not easily summarized by median/mean.
Indeed, we previously found this heterogeneity may be important for influenza evolution: elifesciences.org/reviewed-pre...
Indeed, we previously found this heterogeneity may be important for influenza evolution: elifesciences.org/reviewed-pre...

September 8, 2025 at 9:53 PM
Above visualizations just scratch surface of data: there is tremendous heterogeneity across sera from different individuals not easily summarized by median/mean.
Indeed, we previously found this heterogeneity may be important for influenza evolution: elifesciences.org/reviewed-pre...
Indeed, we previously found this heterogeneity may be important for influenza evolution: elifesciences.org/reviewed-pre...
We then measured how 188 human sera recently collected at four different sites neutralized all 140 influenza strains in library.
Titers are summarized below; can be examined interactively at jbloomlab.github.io/flu-seqneut-... & jbloomlab.github.io/flu-seqneut-...
Titers are summarized below; can be examined interactively at jbloomlab.github.io/flu-seqneut-... & jbloomlab.github.io/flu-seqneut-...
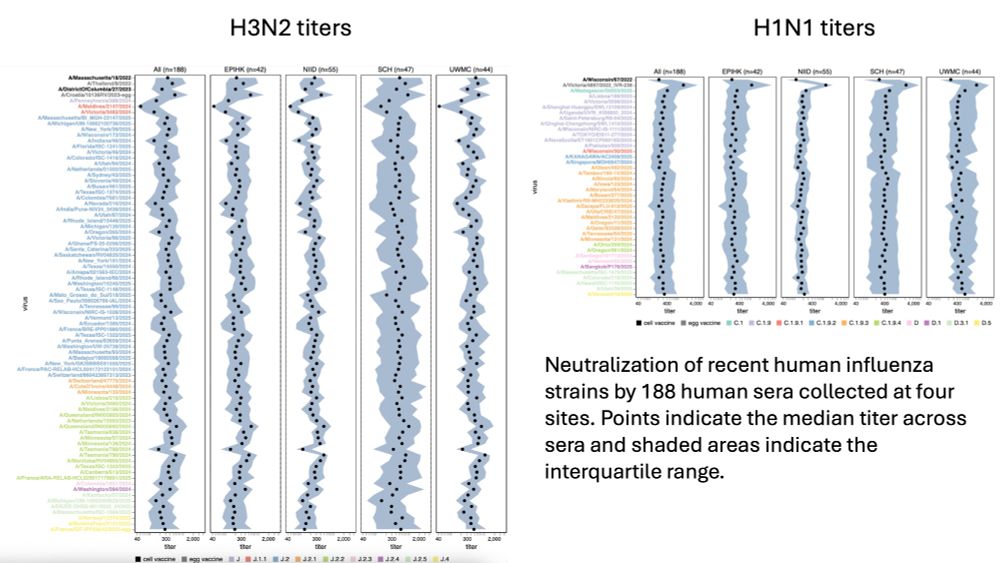
September 8, 2025 at 9:52 PM
We then measured how 188 human sera recently collected at four different sites neutralized all 140 influenza strains in library.
Titers are summarized below; can be examined interactively at jbloomlab.github.io/flu-seqneut-... & jbloomlab.github.io/flu-seqneut-...
Titers are summarized below; can be examined interactively at jbloomlab.github.io/flu-seqneut-... & jbloomlab.github.io/flu-seqneut-...
In spring of 2025, we designed library of naturally occurring human seasonal influenza strains that represented diversity of available sequences at that time; this library continues to cover most sequenced diversity of H3N2 and H1N1 hemagglutinin today.

September 8, 2025 at 9:51 PM
In spring of 2025, we designed library of naturally occurring human seasonal influenza strains that represented diversity of available sequences at that time; this library continues to cover most sequenced diversity of H3N2 and H1N1 hemagglutinin today.
To do this, we used sequencing-based neutralization assays that measure many neutralization curves simultaneously (journals.asm.org/doi/10.1128/... & elifesciences.org/reviewed-pre...)
Approach enabled one grad student (@ckikawa.bsky.social) to measure ~26,000 neutralization curves in ~5 months.
Approach enabled one grad student (@ckikawa.bsky.social) to measure ~26,000 neutralization curves in ~5 months.

September 8, 2025 at 9:51 PM
To do this, we used sequencing-based neutralization assays that measure many neutralization curves simultaneously (journals.asm.org/doi/10.1128/... & elifesciences.org/reviewed-pre...)
Approach enabled one grad student (@ckikawa.bsky.social) to measure ~26,000 neutralization curves in ~5 months.
Approach enabled one grad student (@ckikawa.bsky.social) to measure ~26,000 neutralization curves in ~5 months.
But because it takes time to perform experiments, measurement of how current strains are neutralized by human serum antibodies can lag timeline for vaccine strain selection.
Our goal was to use new approach to characterize human antibody landscape at scale in near real-time.
Our goal was to use new approach to characterize human antibody landscape at scale in near real-time.

September 8, 2025 at 9:49 PM
But because it takes time to perform experiments, measurement of how current strains are neutralized by human serum antibodies can lag timeline for vaccine strain selection.
Our goal was to use new approach to characterize human antibody landscape at scale in near real-time.
Our goal was to use new approach to characterize human antibody landscape at scale in near real-time.
Data in interactive form at dms-vep.org/CHIKV-181-25...
Thanks to Xiaohui Ju for leading study
Special thanks to @msdiamondlab.bsky.social for help
Also Will Hannon, Caelan Radford, Brendan Larsen, Daved Fremont, Ofer Zimmerman, Tomasz Kaszuba, Chris Nelson, Israel Baltazar-Perez, Samantha Nelson
Thanks to Xiaohui Ju for leading study
Special thanks to @msdiamondlab.bsky.social for help
Also Will Hannon, Caelan Radford, Brendan Larsen, Daved Fremont, Ofer Zimmerman, Tomasz Kaszuba, Chris Nelson, Israel Baltazar-Perez, Samantha Nelson

September 4, 2025 at 11:20 PM
Data in interactive form at dms-vep.org/CHIKV-181-25...
Thanks to Xiaohui Ju for leading study
Special thanks to @msdiamondlab.bsky.social for help
Also Will Hannon, Caelan Radford, Brendan Larsen, Daved Fremont, Ofer Zimmerman, Tomasz Kaszuba, Chris Nelson, Israel Baltazar-Perez, Samantha Nelson
Thanks to Xiaohui Ju for leading study
Special thanks to @msdiamondlab.bsky.social for help
Also Will Hannon, Caelan Radford, Brendan Larsen, Daved Fremont, Ofer Zimmerman, Tomasz Kaszuba, Chris Nelson, Israel Baltazar-Perez, Samantha Nelson
After using pseudoviruses & reporter particles to show mutations *loss* of function, we engineered into Chikungunya virus: mutants lost ability to infect human or mosquito cells.
So we reduced natural tropism for both human & mosquito cells to just one type of cell.
So we reduced natural tropism for both human & mosquito cells to just one type of cell.

September 4, 2025 at 11:17 PM
After using pseudoviruses & reporter particles to show mutations *loss* of function, we engineered into Chikungunya virus: mutants lost ability to infect human or mosquito cells.
So we reduced natural tropism for both human & mosquito cells to just one type of cell.
So we reduced natural tropism for both human & mosquito cells to just one type of cell.
We next used non-replicative single-cycle alphavirus reporter particles (which provide another safe way to study mutations) to validate that mutations identified in deep mutational scanning indeed specifically impaired entry in human or mosquito cells only.

September 4, 2025 at 11:16 PM
We next used non-replicative single-cycle alphavirus reporter particles (which provide another safe way to study mutations) to validate that mutations identified in deep mutational scanning indeed specifically impaired entry in human or mosquito cells only.
Sites where mutations specifically impair entry in 293T-MXRA8 cells mostly at MXRA8 binding interface.
We also find sites where mutations specifically impair entry in C6/36 cells. Although mosquito receptor unknown, we hypothesize these sites at its binding interface.
We also find sites where mutations specifically impair entry in C6/36 cells. Although mosquito receptor unknown, we hypothesize these sites at its binding interface.

September 4, 2025 at 11:15 PM
Sites where mutations specifically impair entry in 293T-MXRA8 cells mostly at MXRA8 binding interface.
We also find sites where mutations specifically impair entry in C6/36 cells. Although mosquito receptor unknown, we hypothesize these sites at its binding interface.
We also find sites where mutations specifically impair entry in C6/36 cells. Although mosquito receptor unknown, we hypothesize these sites at its binding interface.
Most mutations similarly affect entry in all three cells, but some have cell-specific effects.
For instance, mutations at E2 site 119 are generally tolerated in C6/36 and 293T-TIM1 cells, but deleterious in 293T-MXRA8 cells.
(See dms-vep.org/CHIKV-181-25... for interactive plot.)
For instance, mutations at E2 site 119 are generally tolerated in C6/36 and 293T-TIM1 cells, but deleterious in 293T-MXRA8 cells.
(See dms-vep.org/CHIKV-181-25... for interactive plot.)

September 4, 2025 at 11:15 PM
Most mutations similarly affect entry in all three cells, but some have cell-specific effects.
For instance, mutations at E2 site 119 are generally tolerated in C6/36 and 293T-TIM1 cells, but deleterious in 293T-MXRA8 cells.
(See dms-vep.org/CHIKV-181-25... for interactive plot.)
For instance, mutations at E2 site 119 are generally tolerated in C6/36 and 293T-TIM1 cells, but deleterious in 293T-MXRA8 cells.
(See dms-vep.org/CHIKV-181-25... for interactive plot.)
We first measured how mutations affect entry in 293T cells expressing human receptor MXRA8.
Below is constraint mapped on structure; see dms-vep.org/CHIKV-181-25... for interactive heatmap of these data.
Below is constraint mapped on structure; see dms-vep.org/CHIKV-181-25... for interactive heatmap of these data.

September 4, 2025 at 11:14 PM
We first measured how mutations affect entry in 293T cells expressing human receptor MXRA8.
Below is constraint mapped on structure; see dms-vep.org/CHIKV-181-25... for interactive heatmap of these data.
Below is constraint mapped on structure; see dms-vep.org/CHIKV-181-25... for interactive heatmap of these data.
We used pseudovirus deep mutational scanning to measure effects of all mutations to envelope proteins in context of single-cycle pseudotyped particles that provide a safe way to study viral protein mutations outside context of fully infectious virus.

September 4, 2025 at 11:13 PM
We used pseudovirus deep mutational scanning to measure effects of all mutations to envelope proteins in context of single-cycle pseudotyped particles that provide a safe way to study viral protein mutations outside context of fully infectious virus.
Chikungunya virus enters cell using its envelope proteins, which are also target of neutralizing antibodies and vaccine design.
A receptor for these viral proteins in mammalian cells is the protein MXRA8, but receptor in mosquito cells is unknown.
A receptor for these viral proteins in mammalian cells is the protein MXRA8, but receptor in mosquito cells is unknown.

September 4, 2025 at 11:13 PM
Chikungunya virus enters cell using its envelope proteins, which are also target of neutralizing antibodies and vaccine design.
A receptor for these viral proteins in mammalian cells is the protein MXRA8, but receptor in mosquito cells is unknown.
A receptor for these viral proteins in mammalian cells is the protein MXRA8, but receptor in mosquito cells is unknown.
As background, Chikungunya virus has transmission cycle that involves infecting both mosquitoes & humans or other primates.
Infection can cause fever and severe joint pain in humans.
Outbreaks are growing due to expanding mosquito range: www.nytimes.com/2025/08/19/h...
Infection can cause fever and severe joint pain in humans.
Outbreaks are growing due to expanding mosquito range: www.nytimes.com/2025/08/19/h...

September 4, 2025 at 11:12 PM
As background, Chikungunya virus has transmission cycle that involves infecting both mosquitoes & humans or other primates.
Infection can cause fever and severe joint pain in humans.
Outbreaks are growing due to expanding mosquito range: www.nytimes.com/2025/08/19/h...
Infection can cause fever and severe joint pain in humans.
Outbreaks are growing due to expanding mosquito range: www.nytimes.com/2025/08/19/h...
Finally, we measured how mutations affect neutralization by three relevant monoclonal antibodies. As shown below, all antibodies adversely affected by mutating site 505 which fortunately remains highly constrained for ACE2 binding. We discuss this interesting site more in preprint.

August 20, 2025 at 5:27 AM
Finally, we measured how mutations affect neutralization by three relevant monoclonal antibodies. As shown below, all antibodies adversely affected by mutating site 505 which fortunately remains highly constrained for ACE2 binding. We discuss this interesting site more in preprint.
We used this fact to estimate how much mutations at each site affect RBD up-down motion, as shown below.
Many of these sites have mutated during SARS-CoV-2 evolution in humans, demonstrating importance of RBD motion and its effects on ACE2 binding & antibody neutralization.
Many of these sites have mutated during SARS-CoV-2 evolution in humans, demonstrating importance of RBD motion and its effects on ACE2 binding & antibody neutralization.

August 20, 2025 at 5:26 AM
We used this fact to estimate how much mutations at each site affect RBD up-down motion, as shown below.
Many of these sites have mutated during SARS-CoV-2 evolution in humans, demonstrating importance of RBD motion and its effects on ACE2 binding & antibody neutralization.
Many of these sites have mutated during SARS-CoV-2 evolution in humans, demonstrating importance of RBD motion and its effects on ACE2 binding & antibody neutralization.
This tradeoff between serum antibody escape and ACE2 binding is because mutations that put the RBD more up enable ACE2 binding but also promote antibody binding. Mutations that put the RBD more down do the opposite.

August 20, 2025 at 5:26 AM
This tradeoff between serum antibody escape and ACE2 binding is because mutations that put the RBD more up enable ACE2 binding but also promote antibody binding. Mutations that put the RBD more down do the opposite.
Some mutations outside the RBD have a strong effect on serum antibody neutralization. But for ACE2-distal or non-RBD mutations, there is a strong tradeoff between serum antibody escape and ACE2 binding as shown below.

August 20, 2025 at 5:25 AM
Some mutations outside the RBD have a strong effect on serum antibody neutralization. But for ACE2-distal or non-RBD mutations, there is a strong tradeoff between serum antibody escape and ACE2 binding as shown below.
Despite imprinting, in some individuals recent infection or vaccination appreciably shift immunodominant neutralizing epitopes.
So new exposures are altering neutralizing serum antibody repertoire, although our data do not define mechanism (see preprint for hypotheses).
So new exposures are altering neutralizing serum antibody repertoire, although our data do not define mechanism (see preprint for hypotheses).

August 20, 2025 at 5:25 AM
Despite imprinting, in some individuals recent infection or vaccination appreciably shift immunodominant neutralizing epitopes.
So new exposures are altering neutralizing serum antibody repertoire, although our data do not define mechanism (see preprint for hypotheses).
So new exposures are altering neutralizing serum antibody repertoire, although our data do not define mechanism (see preprint for hypotheses).
We measured how mutations affected neutralization by sera collected from humans before or after vaccination or infection w recent JN.1-descendant variant. Key sites of escape are shown below; some of the sites (eg, 475 and 478) are mutated in very recent variants.

August 20, 2025 at 5:24 AM
We measured how mutations affected neutralization by sera collected from humans before or after vaccination or infection w recent JN.1-descendant variant. Key sites of escape are shown below; some of the sites (eg, 475 and 478) are mutated in very recent variants.
We next measured how mutations affect full-spike ACE2 binding. Most mutations had similar impacts in KP.3.1.1 and the older XBB.1.5 spike, but some recent mutations (eg, A435S and Q493E) enhance ACE2 binding KP.3.1.1 after impairing it in XBB.1.5.

August 20, 2025 at 5:24 AM
We next measured how mutations affect full-spike ACE2 binding. Most mutations had similar impacts in KP.3.1.1 and the older XBB.1.5 spike, but some recent mutations (eg, A435S and Q493E) enhance ACE2 binding KP.3.1.1 after impairing it in XBB.1.5.
We first measured how all mutations affect cell entry; see this interactive page for those data: dms-vep.org/SARS-CoV-2_K...
A number of mutations that have spread recently are more tolerated in KP.3.1.1 than older XBB.1.5 variant, suggesting epistatic shifts favored their emergence.
A number of mutations that have spread recently are more tolerated in KP.3.1.1 than older XBB.1.5 variant, suggesting epistatic shifts favored their emergence.

August 20, 2025 at 5:23 AM
We first measured how all mutations affect cell entry; see this interactive page for those data: dms-vep.org/SARS-CoV-2_K...
A number of mutations that have spread recently are more tolerated in KP.3.1.1 than older XBB.1.5 variant, suggesting epistatic shifts favored their emergence.
A number of mutations that have spread recently are more tolerated in KP.3.1.1 than older XBB.1.5 variant, suggesting epistatic shifts favored their emergence.
We previously developed pseudovirus deep mutational scanning (pubmed.ncbi.nlm.nih.gov/36868218/), which uses non-replicative viral particles to safely study spike mutations.
Here we used approach to measure how mutations to KP.3.1.1 spike affect five phenotypes, as shown below.
Here we used approach to measure how mutations to KP.3.1.1 spike affect five phenotypes, as shown below.

August 20, 2025 at 5:23 AM
We previously developed pseudovirus deep mutational scanning (pubmed.ncbi.nlm.nih.gov/36868218/), which uses non-replicative viral particles to safely study spike mutations.
Here we used approach to measure how mutations to KP.3.1.1 spike affect five phenotypes, as shown below.
Here we used approach to measure how mutations to KP.3.1.1 spike affect five phenotypes, as shown below.
We examined spike of KP.3.1.1, a strain from late 2024 / early 2025 similar to current variants.
KP.3.1.1 & other recent variants have >60 spike amino-acid mutations relative to early pandemic strains, as spike has evolved at extraordinary rate of >10 mutations/year on average.
KP.3.1.1 & other recent variants have >60 spike amino-acid mutations relative to early pandemic strains, as spike has evolved at extraordinary rate of >10 mutations/year on average.

August 20, 2025 at 5:23 AM
We examined spike of KP.3.1.1, a strain from late 2024 / early 2025 similar to current variants.
KP.3.1.1 & other recent variants have >60 spike amino-acid mutations relative to early pandemic strains, as spike has evolved at extraordinary rate of >10 mutations/year on average.
KP.3.1.1 & other recent variants have >60 spike amino-acid mutations relative to early pandemic strains, as spike has evolved at extraordinary rate of >10 mutations/year on average.
We used pseudovirus deep mutational scanning to look at specificity of neutralization elicited by stabilized vs non-stabilized HA nanoparticle vaccine.
Stabilization directed more neutralization towards potently neutralizing epitopes on top of HA head as shown below.
Stabilization directed more neutralization towards potently neutralizing epitopes on top of HA head as shown below.

August 4, 2025 at 7:16 PM
We used pseudovirus deep mutational scanning to look at specificity of neutralization elicited by stabilized vs non-stabilized HA nanoparticle vaccine.
Stabilization directed more neutralization towards potently neutralizing epitopes on top of HA head as shown below.
Stabilization directed more neutralization towards potently neutralizing epitopes on top of HA head as shown below.

