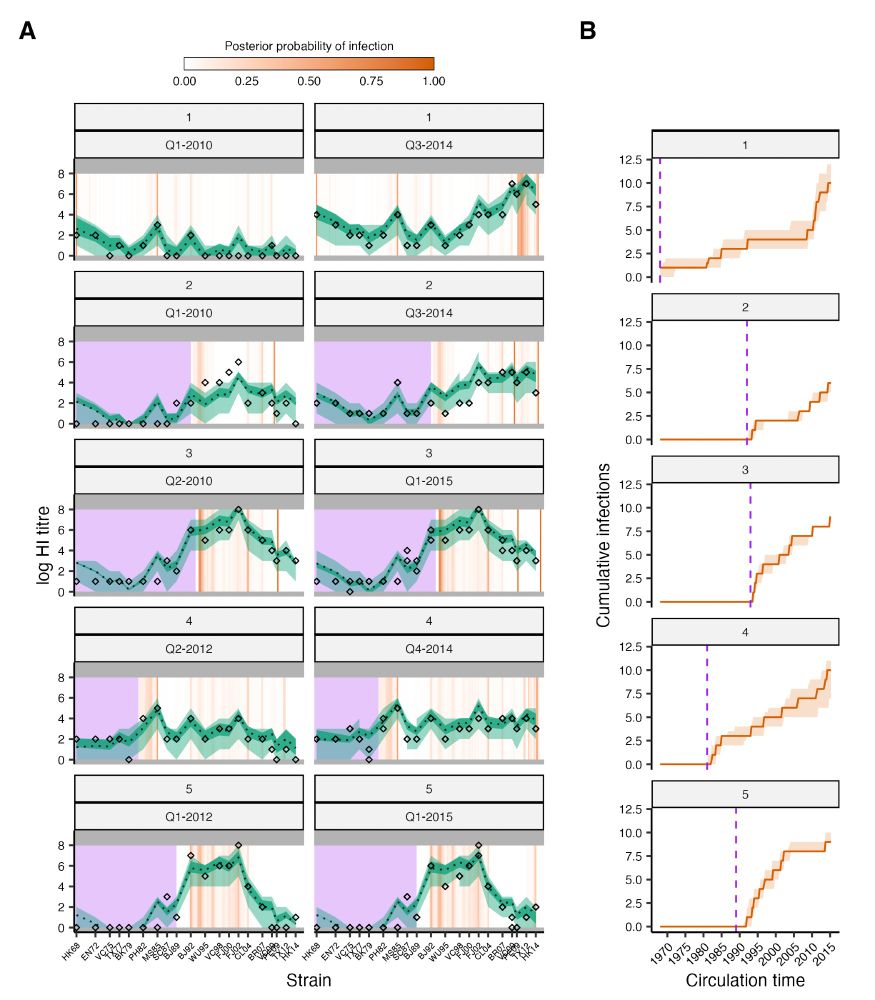
Code and data here: github.com/jameshay218/....
Serosolver package: github.com/seroanalytic...
Code and data here: github.com/jameshay218/....
Serosolver package: github.com/seroanalytic...
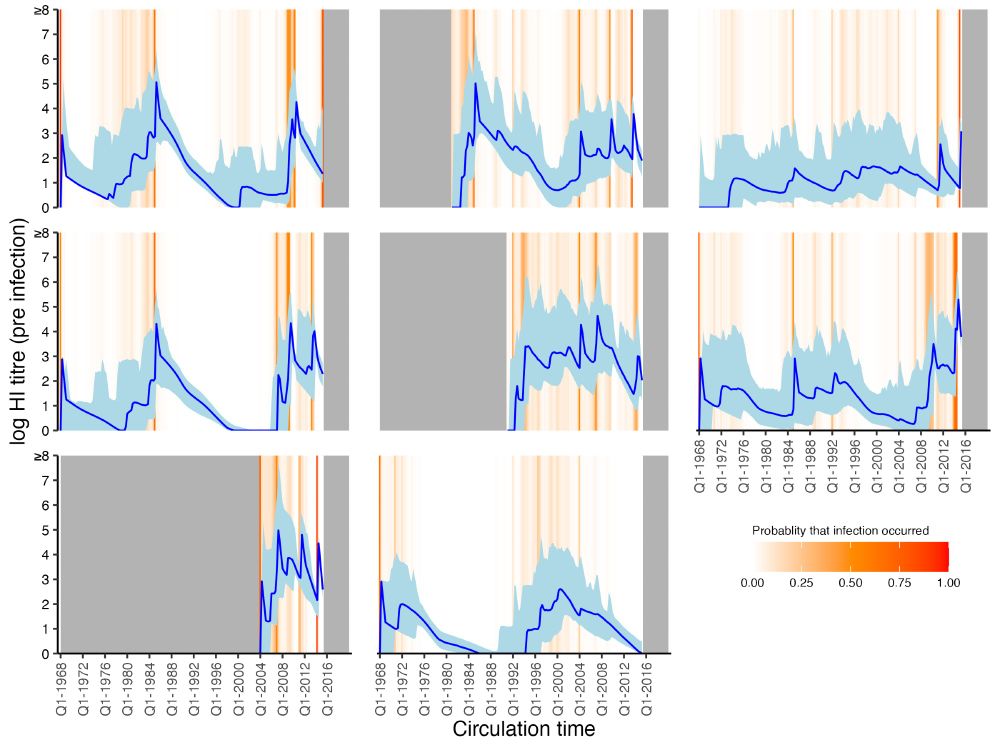
1. Serology-based attack rates are high, at around 18% infected per year.
2. Influenza A/H3N2 infection rates are highest in children, decrease with age and plateau in adulthood.
3. Incidence rates are highly correlated at this small spatial scale.
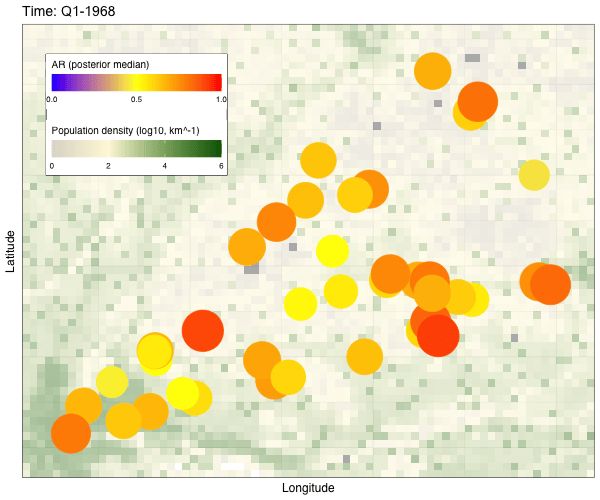
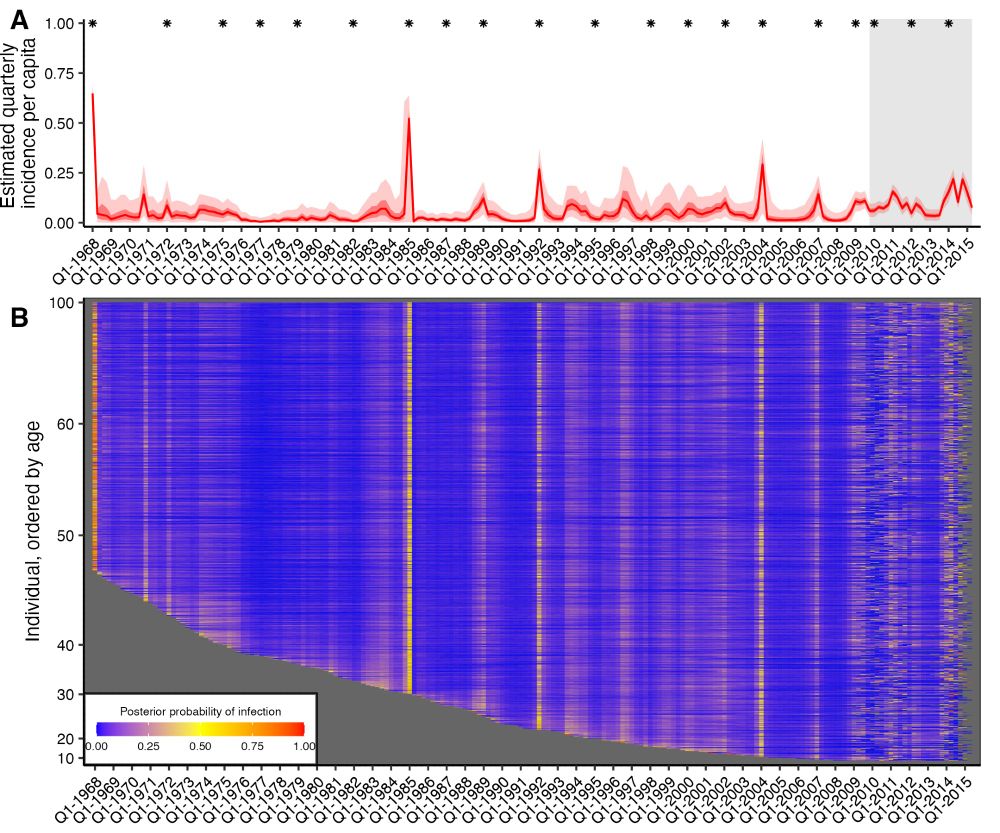
1. Serology-based attack rates are high, at around 18% infected per year.
2. Influenza A/H3N2 infection rates are highest in children, decrease with age and plateau in adulthood.
3. Incidence rates are highly correlated at this small spatial scale.