
Ivan Gushchin
@ivangushchin.bsky.social
Structural biology and protein engineering
"Study of the effect of acoustic treatment on freezing point and boiling point of tap water" - way to go for a scientific journal www.nature.com/articles/s41...

October 29, 2025 at 7:39 PM
"Study of the effect of acoustic treatment on freezing point and boiling point of tap water" - way to go for a scientific journal www.nature.com/articles/s41...
Previously, we engineered 3 artificial CagLOV-derived proteins doi.org/10.1002/pro.... - so I'm glad to see that the approach works for other LOVs as well

October 6, 2025 at 6:23 AM
Previously, we engineered 3 artificial CagLOV-derived proteins doi.org/10.1002/pro.... - so I'm glad to see that the approach works for other LOVs as well
They even succeed in engineering an ultrastable photoactive protein based on Atphot2 LOV2

October 6, 2025 at 6:23 AM
They even succeed in engineering an ultrastable photoactive protein based on Atphot2 LOV2
Interesting preprint by Herzog and colleagues where they investigate 21 diverse LOV domain - also beautiful illustrations throughout

October 6, 2025 at 6:23 AM
Interesting preprint by Herzog and colleagues where they investigate 21 diverse LOV domain - also beautiful illustrations throughout
Glad to contribute to this: in our most recent preprint, we describe peculiar ferritin-like proteins that have ferroxidase activity, but do not form shells, using encapsulin compartments instead: doi.org/10.1101/2025...

October 1, 2025 at 7:36 PM
Glad to contribute to this: in our most recent preprint, we describe peculiar ferritin-like proteins that have ferroxidase activity, but do not form shells, using encapsulin compartments instead: doi.org/10.1101/2025...
And now prohibitins!
www.pnas.org/doi/pdf/10.1...
www.pnas.org/doi/pdf/10.1...

September 27, 2025 at 8:05 PM
And now prohibitins!
www.pnas.org/doi/pdf/10.1...
www.pnas.org/doi/pdf/10.1...
Got an idea while reviewing a grant and a paper
Image courtesy: Qwen
Image courtesy: Qwen

August 11, 2025 at 9:35 AM
Got an idea while reviewing a grant and a paper
Image courtesy: Qwen
Image courtesy: Qwen
Summer holidays @natcomms.nature.com ? 😕

July 21, 2025 at 3:10 PM
Summer holidays @natcomms.nature.com ? 😕
Great visual representation! Looks a little bit like a marketplace: choose the best organism for your specifications

July 20, 2025 at 9:55 AM
Great visual representation! Looks a little bit like a marketplace: choose the best organism for your specifications
Interesting work from my former lab mate Vitaly Polovinkin and colleagues: solving a high resolution structure of a protein inside the cell using electron diffraction

July 12, 2025 at 12:08 PM
Interesting work from my former lab mate Vitaly Polovinkin and colleagues: solving a high resolution structure of a protein inside the cell using electron diffraction
Happy to present a new flavin-binding protein that unexpectedly displays a large Stokes shift due to excited state proton transfer. Not submitted yet - feedback is welcome!
doi.org/10.1101/2025...
doi.org/10.1101/2025...
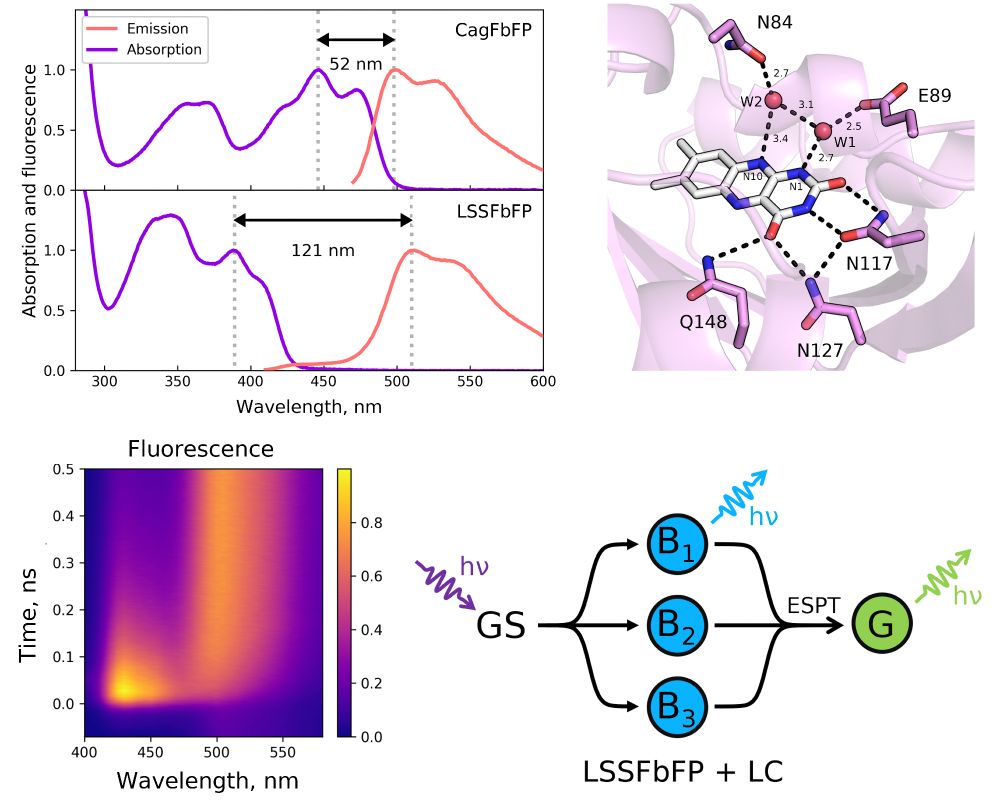
June 17, 2025 at 9:12 AM
Happy to present a new flavin-binding protein that unexpectedly displays a large Stokes shift due to excited state proton transfer. Not submitted yet - feedback is welcome!
doi.org/10.1101/2025...
doi.org/10.1101/2025...
Terrific work from Nikita Egorkin, Nikolai Sluchanko and their colleagues, who discovered a protein binding two carotenoids and two bilins (also four PC lipids!), thus mimicking absorption spectrum of green plants and giving crickets their color. Link to the PNAS article: doi.org/10.1073/pnas...

May 31, 2025 at 12:21 PM
Terrific work from Nikita Egorkin, Nikolai Sluchanko and their colleagues, who discovered a protein binding two carotenoids and two bilins (also four PC lipids!), thus mimicking absorption spectrum of green plants and giving crickets their color. Link to the PNAS article: doi.org/10.1073/pnas...
It looks like Reviewer 1 was happy from the start, Reviewer 2 criticized crystallography and got the paper rejected, but said okay after revision. Still, this data collection and refinement statistics raises quite a number of questions.

May 15, 2025 at 7:36 AM
It looks like Reviewer 1 was happy from the start, Reviewer 2 criticized crystallography and got the paper rejected, but said okay after revision. Still, this data collection and refinement statistics raises quite a number of questions.
Having recently submitted manuscripts to Nature Communications/Science Advances and the like, very surprised to see the data like this published, claiming magnesium interaction. The purported resolution is 2.5 Å
doi.org/10.1038/s414...
doi.org/10.1038/s414...

May 15, 2025 at 7:32 AM
Having recently submitted manuscripts to Nature Communications/Science Advances and the like, very surprised to see the data like this published, claiming magnesium interaction. The purported resolution is 2.5 Å
doi.org/10.1038/s414...
doi.org/10.1038/s414...
This literature/alphafold journey is inspired by Kirill's recent work on flotillin-associated non-rhodopsin (doi.org/10.1101/2025...) first found by Jose M. Haro-Moreno and colleagues (doi.org/10.1128/msys...). AlphaFold kind of sees some transmembrane interaction, but not particularly reliably

May 8, 2025 at 10:27 AM
This literature/alphafold journey is inspired by Kirill's recent work on flotillin-associated non-rhodopsin (doi.org/10.1101/2025...) first found by Jose M. Haro-Moreno and colleagues (doi.org/10.1128/msys...). AlphaFold kind of sees some transmembrane interaction, but not particularly reliably
Interestingly, E. coli flotillin homologue YqiK is so long that it cannot fit in the modeling box (do I understand that correctly?) and bent on itself. Moreover, AF3 is quite sure that it forms a complex with "Inner membrane protein YqiJ"

May 8, 2025 at 10:19 AM
Interestingly, E. coli flotillin homologue YqiK is so long that it cannot fit in the modeling box (do I understand that correctly?) and bent on itself. Moreover, AF3 is quite sure that it forms a complex with "Inner membrane protein YqiJ"
Previous work on bacterial proteins HflK and HflC by Ma and colleagues is also beautiful: they even catch AAA+ proteases inside! This is likely needed to separate proteases from membrane proteins, as shown in the illustration
doi.org/10.1038/s414...
doi.org/10.1038/s414...

May 8, 2025 at 10:14 AM
Previous work on bacterial proteins HflK and HflC by Ma and colleagues is also beautiful: they even catch AAA+ proteases inside! This is likely needed to separate proteases from membrane proteins, as shown in the illustration
doi.org/10.1038/s414...
doi.org/10.1038/s414...
Just learned that it turns out that flotillins form truncated cone-shaped compartments - beautiful structures from Fu and MacKinnon: doi.org/10.1073/pnas...

May 8, 2025 at 10:11 AM
Just learned that it turns out that flotillins form truncated cone-shaped compartments - beautiful structures from Fu and MacKinnon: doi.org/10.1073/pnas...
Also, check out the map quality! Maybe we’ll get to individual protons one day

April 18, 2025 at 7:22 AM
Also, check out the map quality! Maybe we’ll get to individual protons one day
Alternating access mechanism in proton-pumping microbial rhodopsin
Many thanks to everybody involved for the amazing data gathered over the years, and especially to Sergey for getting this over the finish line!
doi.org/10.1126/scia...
Many thanks to everybody involved for the amazing data gathered over the years, and especially to Sergey for getting this over the finish line!
doi.org/10.1126/scia...
April 18, 2025 at 7:18 AM
Alternating access mechanism in proton-pumping microbial rhodopsin
Many thanks to everybody involved for the amazing data gathered over the years, and especially to Sergey for getting this over the finish line!
doi.org/10.1126/scia...
Many thanks to everybody involved for the amazing data gathered over the years, and especially to Sergey for getting this over the finish line!
doi.org/10.1126/scia...
I've tried modeling the effect of double Cys->Ser mutation, it changes the conformation of the sensor, but the transmembrane region remains the same. It is a well known problem of AlphaFold modeling that the conformations of different domains may not match.


March 22, 2025 at 10:17 AM
I've tried modeling the effect of double Cys->Ser mutation, it changes the conformation of the sensor, but the transmembrane region remains the same. It is a well known problem of AlphaFold modeling that the conformations of different domains may not match.
Great work! By the way, AlphaFold3 can make the model with better metrics and linked cysteines, although otherwise it looks very similar to yours. Prediction runs in ~10 minutes golgi.sandbox.google.com


March 22, 2025 at 10:12 AM
Great work! By the way, AlphaFold3 can make the model with better metrics and linked cysteines, although otherwise it looks very similar to yours. Prediction runs in ~10 minutes golgi.sandbox.google.com
We used a related approach to predict stoichiometry of high-order circular homo-oligomers (ATPase c-rings), onlinelibrary.wiley.com/doi/full/10.... - although not so sophisticated, worked well on a test set including proteins, for which only AFM data is available

March 17, 2025 at 5:19 PM
We used a related approach to predict stoichiometry of high-order circular homo-oligomers (ATPase c-rings), onlinelibrary.wiley.com/doi/full/10.... - although not so sophisticated, worked well on a test set including proteins, for which only AFM data is available
We've tried this with spinach chloroplast ATPase rotor ring, which should be a tetradecamer (14-mer), and everything from 10 to 21 was modeled as a ring by AF2 doi.org/10.1002/prot...

January 26, 2025 at 11:11 PM
We've tried this with spinach chloroplast ATPase rotor ring, which should be a tetradecamer (14-mer), and everything from 10 to 21 was modeled as a ring by AF2 doi.org/10.1002/prot...

