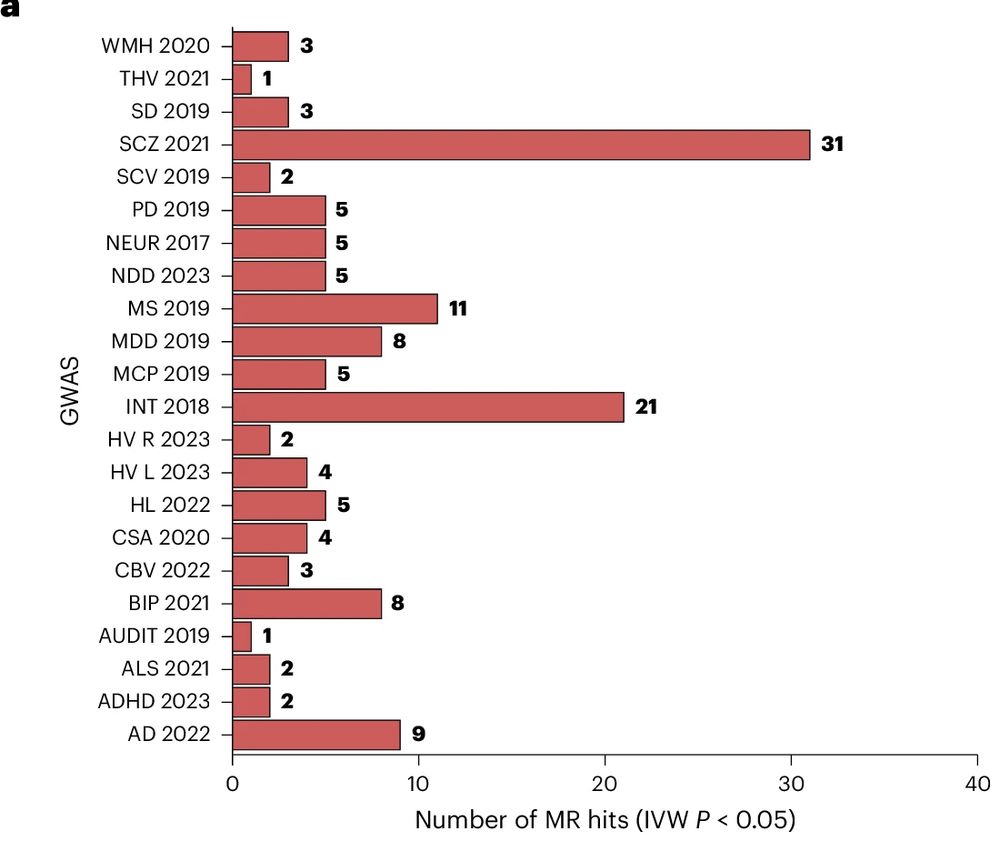
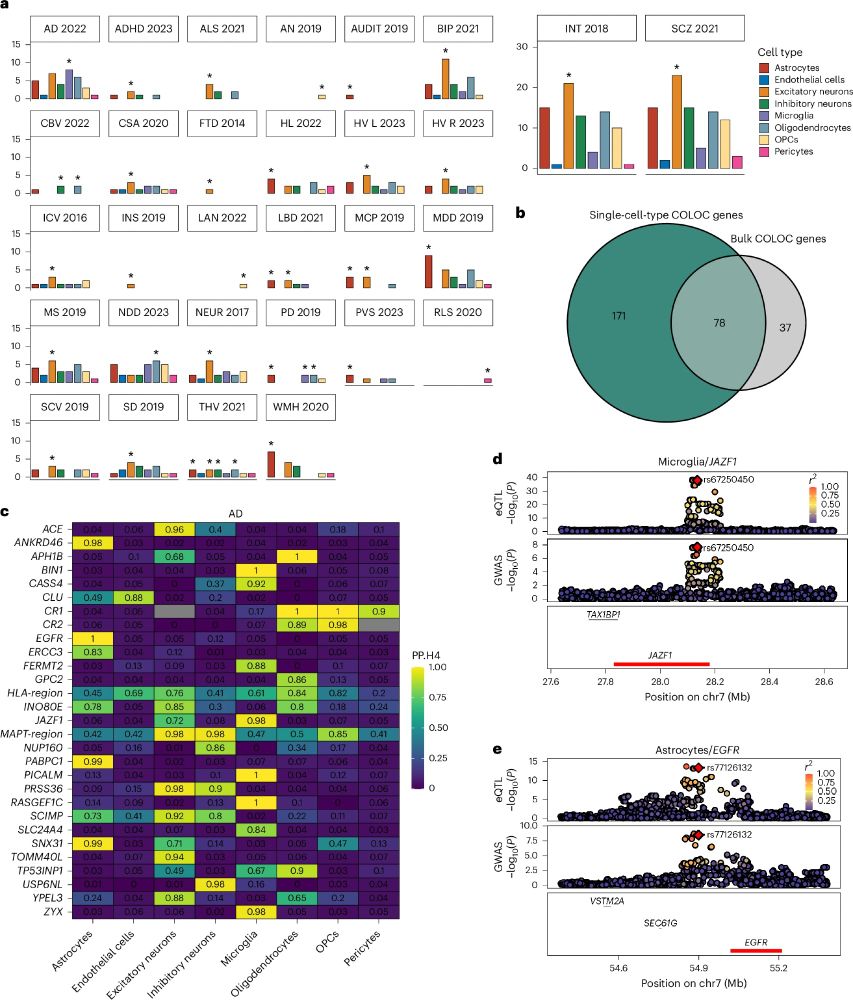
The Landmark project is a public-private partnership involving GSK, Novartis, Roche and UCB with the hope to find novel targets for Parkinson’s Disease.
www.imperial.ac.uk/news/256397/...
Watch this space! 😄
The Landmark project is a public-private partnership involving GSK, Novartis, Roche and UCB with the hope to find novel targets for Parkinson’s Disease.
www.imperial.ac.uk/news/256397/...
Watch this space! 😄
www.nature.com/articles/s41....
If you are interested to hear more, I will be presenting this work virtually at #ADPD2025 if you feel like dropping in!
www.nature.com/articles/s41....
If you are interested to hear more, I will be presenting this work virtually at #ADPD2025 if you feel like dropping in!