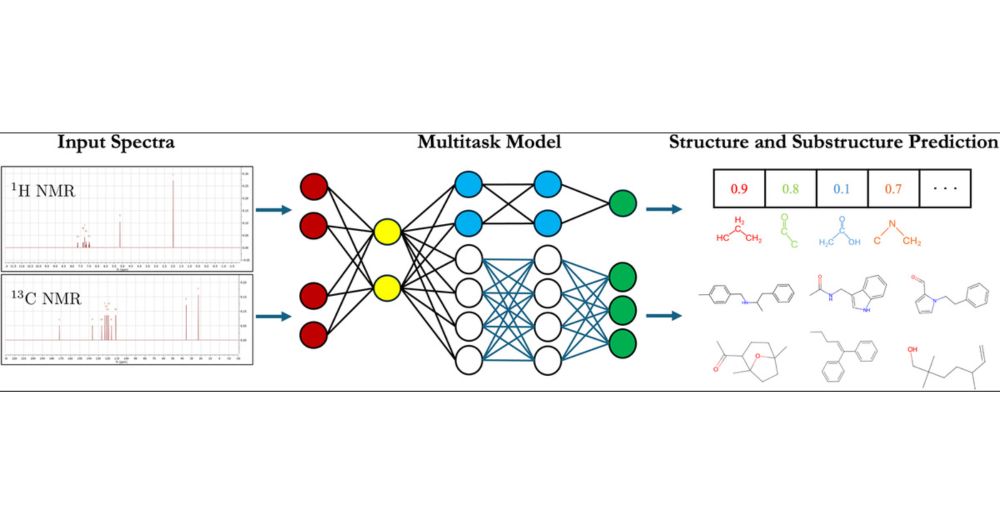
"Characterizing structural and kinetic ensembles of intrinsically disordered proteins using writhe"
www.biorxiv.org/content/10.1...
by Tommy Sisk, with a generative modeling component done in collaboration with @smnlssn.bsky.social
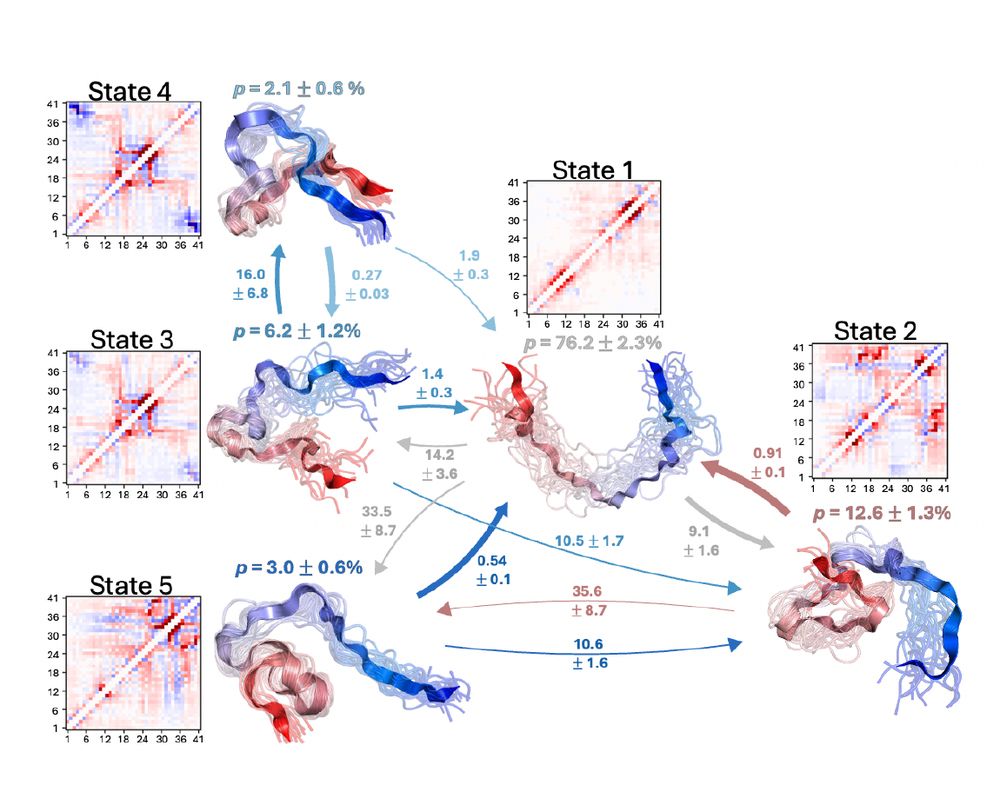
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
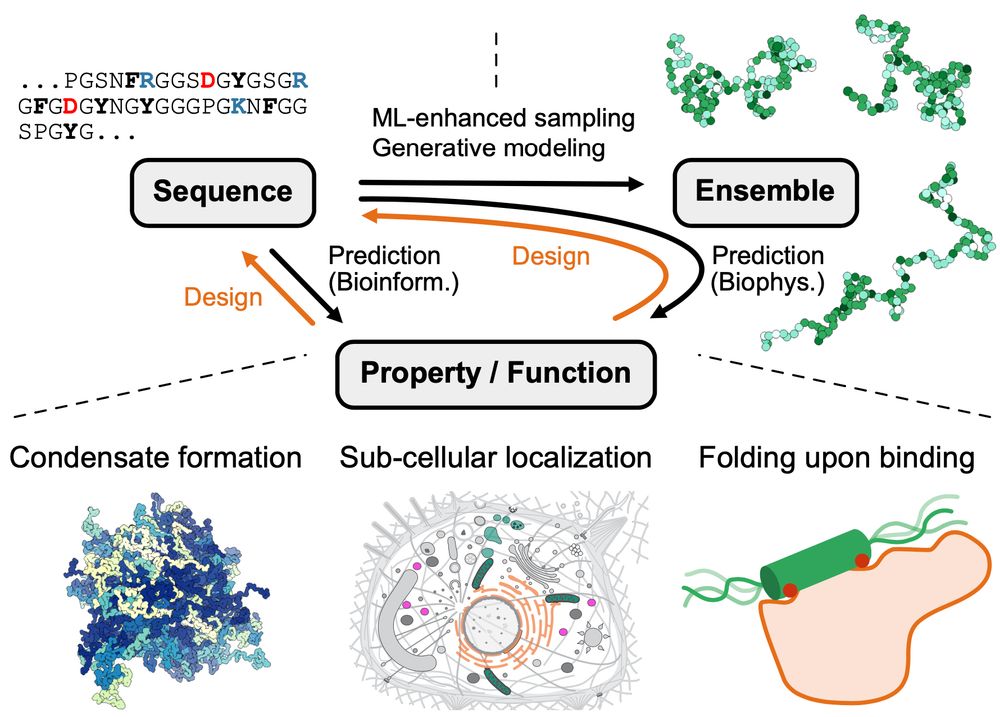
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
Details here: www.futurehouse.org/fellowship

Details here: www.futurehouse.org/fellowship
To me, the most important are:
Read often, read broadly (incl. older papers and outside your field), and learn to read some papers in detail and others more superficially (and quickly)

To me, the most important are:
Read often, read broadly (incl. older papers and outside your field), and learn to read some papers in detail and others more superficially (and quickly)
To celebrate the publication of our data extraction tutorial in Chem Soc Rev, we made it easy to run it — without any installation — on a JupyterHub of the Base4NFDI.
🎥 Video intro to the JupyterHub deployment: youtu.be/l-5QNUo1fcU



If in CADD, pls read through D3R's last paper
pubmed.ncbi.nlm.nih.gov/31974851/

If in CADD, pls read through D3R's last paper
pubmed.ncbi.nlm.nih.gov/31974851/
www.biorxiv.org/content/10.1...