
Genome Informatics & Phylogenetics | giphy.pasteur.fr | CRBIP | crbip.pasteur.fr | Institut Pasteur
#phylogenetics #systematics #taxonomy #genomics #bioinformatics #algorithmics
✅ new confidence score for each inferred #long-read alien oligos
✅ ability to filter out #long-read alien oligos with low score
✅ still fast
gitlab.pasteur.fr/GIPhy/AlienDiscover

✅ new confidence score for each inferred #long-read alien oligos
✅ ability to filter out #long-read alien oligos with low score
✅ still fast
gitlab.pasteur.fr/GIPhy/AlienDiscover
#bioinformatics #FASTQ #16S #rRNA
➡️ new preset options for better sensitivity 🧬
➡️ up-to-date SSU reference databank 🦠
gitlab.pasteur.fr/GIPhy/ASSU
research.pasteur.fr/en/tool/assu

#bioinformatics #FASTQ #16S #rRNA
➡️ new preset options for better sensitivity 🧬
➡️ up-to-date SSU reference databank 🦠
gitlab.pasteur.fr/GIPhy/ASSU
research.pasteur.fr/en/tool/assu

@pasteur.fr 🚼 🌍 🌏
research.pasteur.fr/en/b/1AV1
dx.doi.org/10.1038/s41467-025-65352-4

@pasteur.fr 🚼 🌍 🌏
research.pasteur.fr/en/b/1AV1
dx.doi.org/10.1038/s41467-025-65352-4
The 2nd Nanopore Users Day will take place 🗓️10 March 2026 at the Duclaux Amphitheater, Institut Pasteur (Paris). This edition will be open to external participants!
👉 More details and registration info coming soon!
@chloebaum.bsky.social @pasteur.fr @nanoporetech.com

The 2nd Nanopore Users Day will take place 🗓️10 March 2026 at the Duclaux Amphitheater, Institut Pasteur (Paris). This edition will be open to external participants!
👉 More details and registration info coming soon!
@chloebaum.bsky.social @pasteur.fr @nanoporetech.com
gitlab.pasteur.fr/GIPhy/MSTclust
gitlab.pasteur.fr/GIPhy/MSTclust
#phylogenetics #genomics #taxonomy
@pasteur.fr @microbiologysociety.org
research.pasteur.fr/en/b/18sC
dx.doi.org/10.1099/ijsem.0.006869

#phylogenetics #genomics #taxonomy
@pasteur.fr @microbiologysociety.org
research.pasteur.fr/en/b/18sC
dx.doi.org/10.1099/ijsem.0.006869
#bioinformatics #short-reads #long-reads
gitlab.pasteur.fr/GIPhy/FQsum

#bioinformatics #short-reads #long-reads
gitlab.pasteur.fr/GIPhy/FQsum
inference of alien oligonucleotides (adapter, primer, index, barcode, ...) without any prior nor external knowledge
➡️ still accurate on #short-read #FASTQ files
➡️ now able to deal with #long-read #FASTQ files
➡️ very fast
gitlab.pasteur.fr/GIPhy/AlienDiscover

inference of alien oligonucleotides (adapter, primer, index, barcode, ...) without any prior nor external knowledge
➡️ still accurate on #short-read #FASTQ files
➡️ now able to deal with #long-read #FASTQ files
➡️ very fast
gitlab.pasteur.fr/GIPhy/AlienDiscover
@sylvainbrisse.bsky.social @pasteur.fr
☑️ now able to clip low-residue content regions
☑️ deal with Phred scores up to 95
☑️ still one of the fastest #FASTQ clipping/trimming tool
gitlab.pasteur.fr/GIPhy/AlienTrimmer
research.pasteur.fr/en/software/alientrimmer

@sylvainbrisse.bsky.social @pasteur.fr
☑️ now able to clip low-residue content regions
☑️ deal with Phred scores up to 95
☑️ still one of the fastest #FASTQ clipping/trimming tool
gitlab.pasteur.fr/GIPhy/AlienTrimmer
research.pasteur.fr/en/software/alientrimmer
#bioinformatics #FASTQ #LongReads
➡️ now able to quickly perform digital normalization on long-read data files
gitlab.pasteur.fr/vlegrand/ROCK
research.pasteur.fr/en/software/rock

#bioinformatics #FASTQ #LongReads
➡️ now able to quickly perform digital normalization on long-read data files
gitlab.pasteur.fr/vlegrand/ROCK
research.pasteur.fr/en/software/rock
multi-threaded #FASTQ file downloading from the European Nucleotide Archive (ENA) repository
➡️ now deals with the last ENA file report format
gitlab.pasteur.fr/GIPhy/wgetENAHTS

multi-threaded #FASTQ file downloading from the European Nucleotide Archive (ENA) repository
➡️ now deals with the last ENA file report format
gitlab.pasteur.fr/GIPhy/wgetENAHTS


www.nature.com/articles/s41467-025-60065-0
Congratulations to @chcrestani.bsky.social and @sylvainbrisse.bsky.social 👍

www.nature.com/articles/s41467-025-60065-0
Congratulations to @chcrestani.bsky.social and @sylvainbrisse.bsky.social 👍
#genomics #FASTQ #bioinformatics
✔️ new accuracy index
✔️ compatible with up-to-date dependency versions
✔️ several bugs fixed
gitlab.pasteur.fr/GIPhy/fq2dna
research.pasteur.fr/en/tool/fq2dna

#genomics #FASTQ #bioinformatics
✔️ new accuracy index
✔️ compatible with up-to-date dependency versions
✔️ several bugs fixed
gitlab.pasteur.fr/GIPhy/fq2dna
research.pasteur.fr/en/tool/fq2dna
www.nature.com/articles/s43588-025-00784-y
www.nature.com/articles/s43588-025-00784-y
#bioinformatics #FASTQ
➡️ updated list of alien oligos
➡️ updated trap and TMPDIR handling
➡️ several fixed minor bugs
gitlab.pasteur.fr/GIPhy/fqCleanER
research.pasteur.fr/en/tool/fqcleaner

#bioinformatics #FASTQ
➡️ updated list of alien oligos
➡️ updated trap and TMPDIR handling
➡️ several fixed minor bugs
gitlab.pasteur.fr/GIPhy/fqCleanER
research.pasteur.fr/en/tool/fqcleaner
www.springernature.com/gp/open-scie...
www.springernature.com/gp/open-scie...
#taxonomy #phylogenetics #genomics
www.microbiologyresearch.org/content/jour...

#taxonomy #phylogenetics #genomics
www.microbiologyresearch.org/content/jour...
#bioinformatics #FASTQ
=> fixed bug that (rarely) occurs when processing very large FASTQ files
gitlab.pasteur.fr/vlegrand/ROCK
research.pasteur.fr/en/software/rock

#bioinformatics #FASTQ
=> fixed bug that (rarely) occurs when processing very large FASTQ files
gitlab.pasteur.fr/vlegrand/ROCK
research.pasteur.fr/en/software/rock
Les soumissions pour #JOBIM2025 sont ouvertes ! 😱
💡Articles longs, posters, démos, etc ; de nombreux formats sont attendus.
Les consignes détailles seront accessibles sur le site de #JOBIM2025 ➡️ jobim2025.labri.fr
⏳ Les soumissions fermeront le 31 mars, alors faîtes vite !
Les soumissions pour #JOBIM2025 sont ouvertes ! 😱
💡Articles longs, posters, démos, etc ; de nombreux formats sont attendus.
Les consignes détailles seront accessibles sur le site de #JOBIM2025 ➡️ jobim2025.labri.fr
⏳ Les soumissions fermeront le 31 mars, alors faîtes vite !
link.springer.com/article/10.1...

link.springer.com/article/10.1...

doi.org/10.1093/jac/...
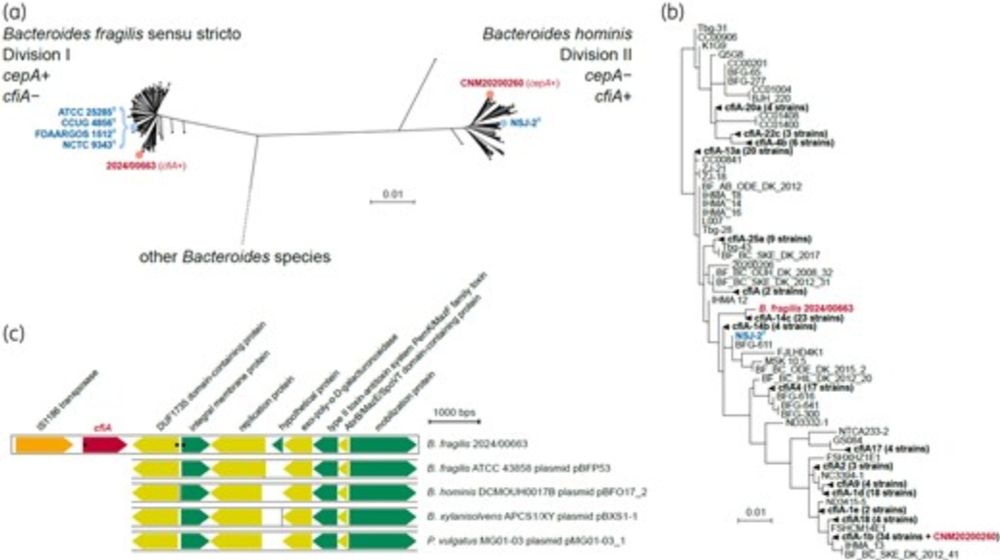
doi.org/10.1093/jac/...

