Hanbin Lee
@epigenci.bsky.social
PhD Student at UMich Statistics.
The account mostly trashes about urban planning and infrastructure.
Probability, Statistics, and Evolutionary Biology.
https://hanbin973.github.io
The account mostly trashes about urban planning and infrastructure.
Probability, Statistics, and Evolutionary Biology.
https://hanbin973.github.io
Why do people with college degrees or higher can't hold themselves from saying these disrespectful words against activities they think aren't as cool as their own jobs?
If you seriously think doing research is any cooler than selling clothoes, you don't deverse the taxes paid by small businesses.
If you seriously think doing research is any cooler than selling clothoes, you don't deverse the taxes paid by small businesses.

September 14, 2025 at 3:29 PM
Why do people with college degrees or higher can't hold themselves from saying these disrespectful words against activities they think aren't as cool as their own jobs?
If you seriously think doing research is any cooler than selling clothoes, you don't deverse the taxes paid by small businesses.
If you seriously think doing research is any cooler than selling clothoes, you don't deverse the taxes paid by small businesses.
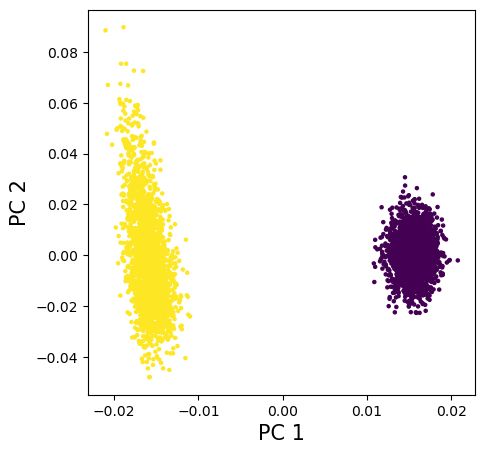
In several spatial simulations, genetic distance based on inferred ARGs are only off by a constant from the true ARG. Subsequently, the PCs are the same for inferred and true ARGs despite the inaccuracy of tree inference at a finer scale (as you see, the ages of individual nodes are quite noisy).

July 20, 2025 at 9:16 AM
In several spatial simulations, genetic distance based on inferred ARGs are only off by a constant from the true ARG. Subsequently, the PCs are the same for inferred and true ARGs despite the inaccuracy of tree inference at a finer scale (as you see, the ages of individual nodes are quite noisy).
This is from my personal website. This time, the prior was driven by mutation. More examples coming soon.

July 19, 2025 at 10:09 AM
This is from my personal website. This time, the prior was driven by mutation. More examples coming soon.
Surprisingly, branch genetic relatedness derived from inferred trees are usually off only by a constant from the true value. Hence, the mixed model with inferred trees does just as good as the true ARG, as shown demonstrated by a spatial simulation.

July 19, 2025 at 2:09 AM
Surprisingly, branch genetic relatedness derived from inferred trees are usually off only by a constant from the true value. Hence, the mixed model with inferred trees does just as good as the true ARG, as shown demonstrated by a spatial simulation.
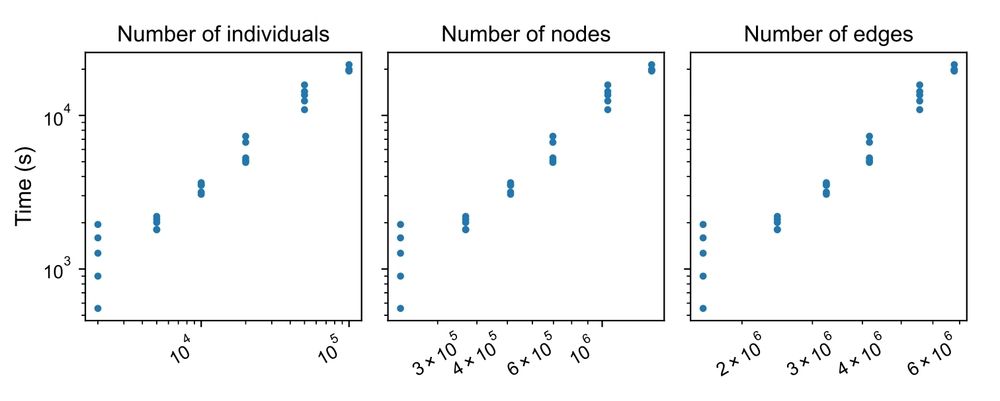
The runtime is linear w.r.t. sample size. How? we used the new matvec algorithm presented in an earlier preprint by Brieuc Lehmann.
I designed a second order Newton update that is almost as cheap as a first order gradient descent.
www.biorxiv.org/content/10.1...
I designed a second order Newton update that is almost as cheap as a first order gradient descent.
www.biorxiv.org/content/10.1...
July 19, 2025 at 2:06 AM
The runtime is linear w.r.t. sample size. How? we used the new matvec algorithm presented in an earlier preprint by Brieuc Lehmann.
I designed a second order Newton update that is almost as cheap as a first order gradient descent.
www.biorxiv.org/content/10.1...
I designed a second order Newton update that is almost as cheap as a first order gradient descent.
www.biorxiv.org/content/10.1...
(3) Where does genetic confounding come from? We show that fixed effects covariates don't exist when the evolution is neutral: YOU DONT NEED TO ADJUST FOR PCs if you think the evolution was indeed neutral (although we know it's not true in practice).

July 19, 2025 at 2:00 AM
(3) Where does genetic confounding come from? We show that fixed effects covariates don't exist when the evolution is neutral: YOU DONT NEED TO ADJUST FOR PCs if you think the evolution was indeed neutral (although we know it's not true in practice).
(2) Pseudoreplication is inevitable in estimating genetic signals. Even if the two cohorts are disjoint, they still have a shared ancestry that leads to dependent parameter estimates. The following is an example of variance component estimates from two non overlapping samples.

July 19, 2025 at 1:56 AM
(2) Pseudoreplication is inevitable in estimating genetic signals. Even if the two cohorts are disjoint, they still have a shared ancestry that leads to dependent parameter estimates. The following is an example of variance component estimates from two non overlapping samples.
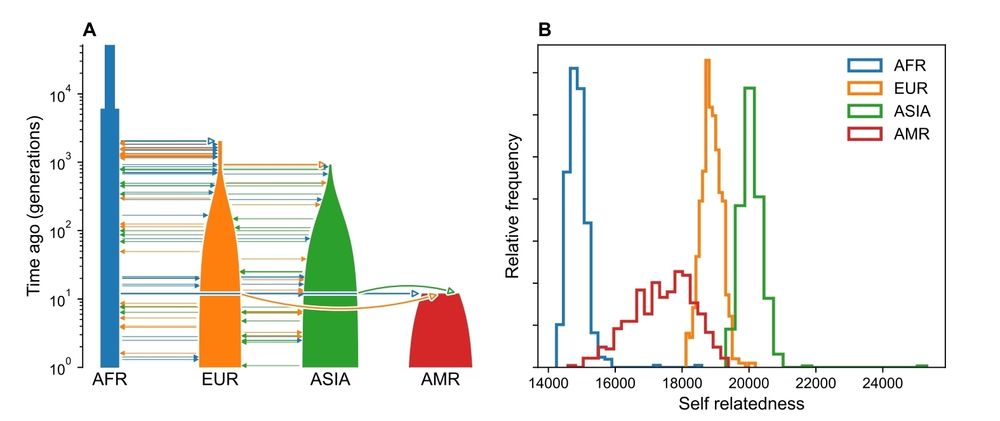
So what do we learn from the theoretical argument?
(1) The underlying demography governs the upper bound of polygenic prediction accuracy.
Why? more inbred/bottlenecked populations have higher genetic variance.
(1) The underlying demography governs the upper bound of polygenic prediction accuracy.
Why? more inbred/bottlenecked populations have higher genetic variance.
July 19, 2025 at 1:54 AM
So what do we learn from the theoretical argument?
(1) The underlying demography governs the upper bound of polygenic prediction accuracy.
Why? more inbred/bottlenecked populations have higher genetic variance.
(1) The underlying demography governs the upper bound of polygenic prediction accuracy.
Why? more inbred/bottlenecked populations have higher genetic variance.
In real data, one could expect that if the Tractor and single-continental marginal effects are the same, running LDSC on the these marginal effect size summary statistics would yield a high genetic correlation. This also seems wrong (some caveats of the analysis in the paper)

June 13, 2025 at 1:53 PM
In real data, one could expect that if the Tractor and single-continental marginal effects are the same, running LDSC on the these marginal effect size summary statistics would yield a high genetic correlation. This also seems wrong (some caveats of the analysis in the paper)
If the assumption holds, one can show that Tractor and single-continental GWAS marginal effect sizes should be equal. This is not true in simulations even if we test the true causal loci.

June 13, 2025 at 1:49 PM
If the assumption holds, one can show that Tractor and single-continental GWAS marginal effect sizes should be equal. This is not true in simulations even if we test the true causal loci.
The part I like the most in the upcoming paper and something that is commonly overlooked in statistical genetics methods development. We too often try to combine two results from drastically different views.

June 11, 2025 at 6:20 AM
The part I like the most in the upcoming paper and something that is commonly overlooked in statistical genetics methods development. We too often try to combine two results from drastically different views.
We demonstrate the new software using a pedigree of UK Labradors consisting of roughly 1.5 million observations.
(2/n)
(2/n)

March 19, 2025 at 12:55 PM
We demonstrate the new software using a pedigree of UK Labradors consisting of roughly 1.5 million observations.
(2/n)
(2/n)
Is this research federally funded?

March 14, 2025 at 7:27 PM
Is this research federally funded?
The Parliament passed the impeachment!

December 14, 2024 at 12:56 PM
The Parliament passed the impeachment!
Dua Lipa is continuing her Korea Tour regardless. She's a pro.

December 4, 2024 at 6:59 AM
Dua Lipa is continuing her Korea Tour regardless. She's a pro.
It is officially a self coup now.

December 3, 2024 at 2:35 PM
It is officially a self coup now.
My vacation plan is about to turn into a...

December 3, 2024 at 2:01 PM
My vacation plan is about to turn into a...
I now have a quantitative measure of the amount of work I have done.

November 16, 2024 at 5:11 PM
I now have a quantitative measure of the amount of work I have done.
My poster at the KOGO conference. Interpreting SNPs as branches gives many neat result across GWAS applications. Most of all, I like the part proving that genomic random effects model is a simple consequence of mutation on ARGs.

February 1, 2024 at 2:40 PM
My poster at the KOGO conference. Interpreting SNPs as branches gives many neat result across GWAS applications. Most of all, I like the part proving that genomic random effects model is a simple consequence of mutation on ARGs.
implemented randomized SVD with tskit.
the randomized algorithm is ~x100 faster with ~1000 samples
the randomized algorithm is ~x100 faster with ~1000 samples

January 8, 2024 at 4:30 AM
implemented randomized SVD with tskit.
the randomized algorithm is ~x100 faster with ~1000 samples
the randomized algorithm is ~x100 faster with ~1000 samples
This paper by Henderson is super cool. It utilizes the statistical interpretation of Cholesky decomposition to obtain a sparse representation of a kinship matrix.

December 7, 2023 at 2:11 PM
This paper by Henderson is super cool. It utilizes the statistical interpretation of Cholesky decomposition to obtain a sparse representation of a kinship matrix.
recombination is indeed tricky
I think current reference-based analyses will become less popular soon.
I think current reference-based analyses will become less popular soon.

November 15, 2023 at 1:58 PM
recombination is indeed tricky
I think current reference-based analyses will become less popular soon.
I think current reference-based analyses will become less popular soon.
Cell-wise methods are the correct choice in the second scenario, like perturb-seq experiments.
The true variance and cell-wise methods' variance estimates are on the same scale.
This is concordant with the simulations.
The true variance and cell-wise methods' variance estimates are on the same scale.
This is concordant with the simulations.

November 6, 2023 at 12:28 PM
Cell-wise methods are the correct choice in the second scenario, like perturb-seq experiments.
The true variance and cell-wise methods' variance estimates are on the same scale.
This is concordant with the simulations.
The true variance and cell-wise methods' variance estimates are on the same scale.
This is concordant with the simulations.