
Ed Chuong
@edchuong.bsky.social
Assistant Professor at the University of Colorado Boulder - Genome regulation, Transposons, Immunity - https://chuonglab.colorado.edu
Reposted by Ed Chuong
I created a brief spreadsheet of reductions I've heard of so far. Any additions you know of (especially if you have the links/receipts) would be great: docs.google.com/spreadsheets...
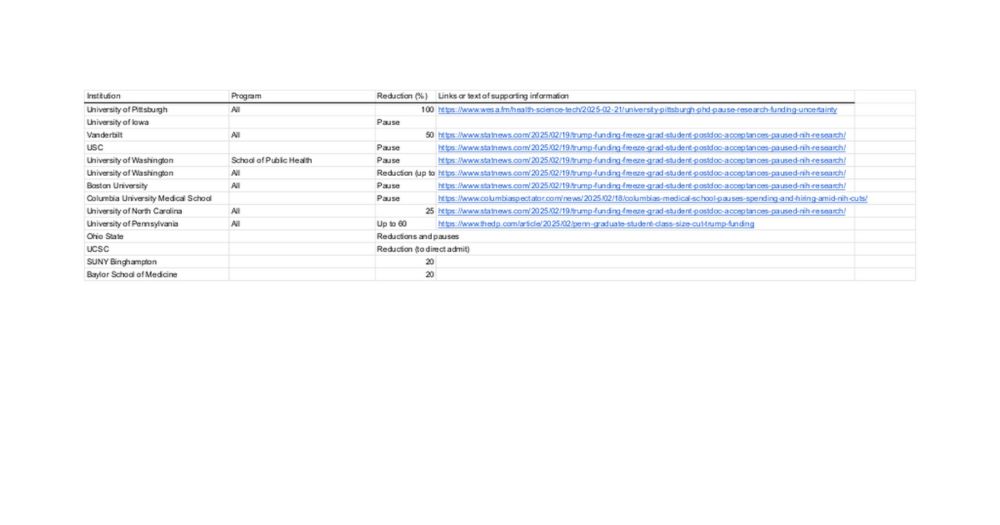
Graduate Reductions Across Biomedical Sciences (2025)
docs.google.com
February 22, 2025 at 1:23 PM
I created a brief spreadsheet of reductions I've heard of so far. Any additions you know of (especially if you have the links/receipts) would be great: docs.google.com/spreadsheets...
Congrats to the team, this is super important for studying TEs!
February 5, 2025 at 7:43 PM
Congrats to the team, this is super important for studying TEs!
Awesome, congrats to the team! 🎉
January 10, 2025 at 4:37 PM
Awesome, congrats to the team! 🎉
Also in this same issue--a fantastic deep dive into the prevalence of transposon exonization by @yarribas.bsky.social !
www.cell.com/cell/fulltex...
www.cell.com/cell/fulltex...

Transposable element exonization generates a reservoir of evolving and functional protein isoforms
Transposable element exonization by unannotated splicing events produces stable protein isoforms with acquired functions that are subject to evolutionary selection.
www.cell.com
December 12, 2024 at 8:31 PM
Also in this same issue--a fantastic deep dive into the prevalence of transposon exonization by @yarribas.bsky.social !
www.cell.com/cell/fulltex...
www.cell.com/cell/fulltex...
All the credit for this discovery goes to postdoc @giuliapasquesi.bsky.social , who first noticed the isoform in her 2020 Lockdown Sideproject (tm). Her perseverance, creativity, and talent made this study possible and it was an honor to be a part of it. Watch out for her--she's on the job market! 😉

December 12, 2024 at 6:57 PM
All the credit for this discovery goes to postdoc @giuliapasquesi.bsky.social , who first noticed the isoform in her 2020 Lockdown Sideproject (tm). Her perseverance, creativity, and talent made this study possible and it was an honor to be a part of it. Watch out for her--she's on the job market! 😉
Beyond IFNAR2, our findings suggest that TE exonization may be a widespread yet hidden source of decoy isoforms that regulate immune signaling.

December 12, 2024 at 6:57 PM
Beyond IFNAR2, our findings suggest that TE exonization may be a widespread yet hidden source of decoy isoforms that regulate immune signaling.
Our study shows how TE exonization gave rise to a primate-specific IFN decoy receptor, which acts as a dial to turn down IFN signaling in human cells. While IFN decoy receptors have evolved repeatedly in viruses (eg VACV B18), this is the first host-encoded IFN decoy receptor.

December 12, 2024 at 6:57 PM
Our study shows how TE exonization gave rise to a primate-specific IFN decoy receptor, which acts as a dial to turn down IFN signaling in human cells. While IFN decoy receptors have evolved repeatedly in viruses (eg VACV B18), this is the first host-encoded IFN decoy receptor.
We also found that risk variants linked to severe COVID-19 in the IFNAR2 locus almost perfectly coincided with splicing QTLs associated with higher relative expression of the decoy. This suggests that variation in IFNAR2 splicing partly underlies individual variation in response to infection.

December 12, 2024 at 6:57 PM
We also found that risk variants linked to severe COVID-19 in the IFNAR2 locus almost perfectly coincided with splicing QTLs associated with higher relative expression of the decoy. This suggests that variation in IFNAR2 splicing partly underlies individual variation in response to infection.

