📍Heidelberg
🦠Human microbiome | Host-microbiome interactions | Multiomics integration
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
First time to cover new ETE4 features: Interactive viz of huge tree, profiling...
In Madrid, April 21-25. Travel fellowships available!

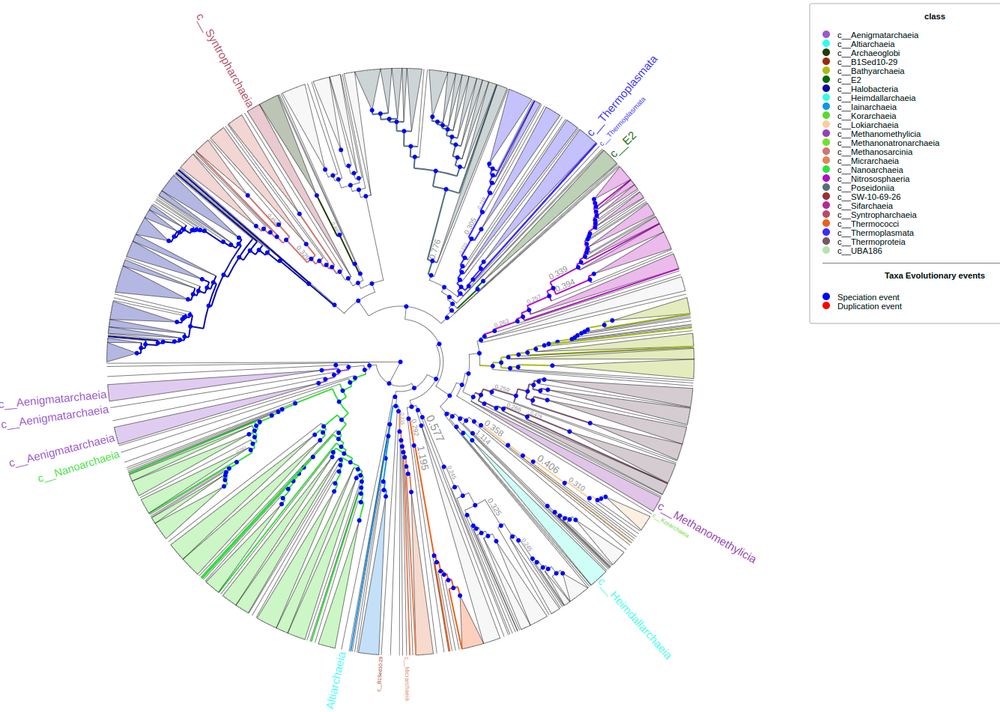
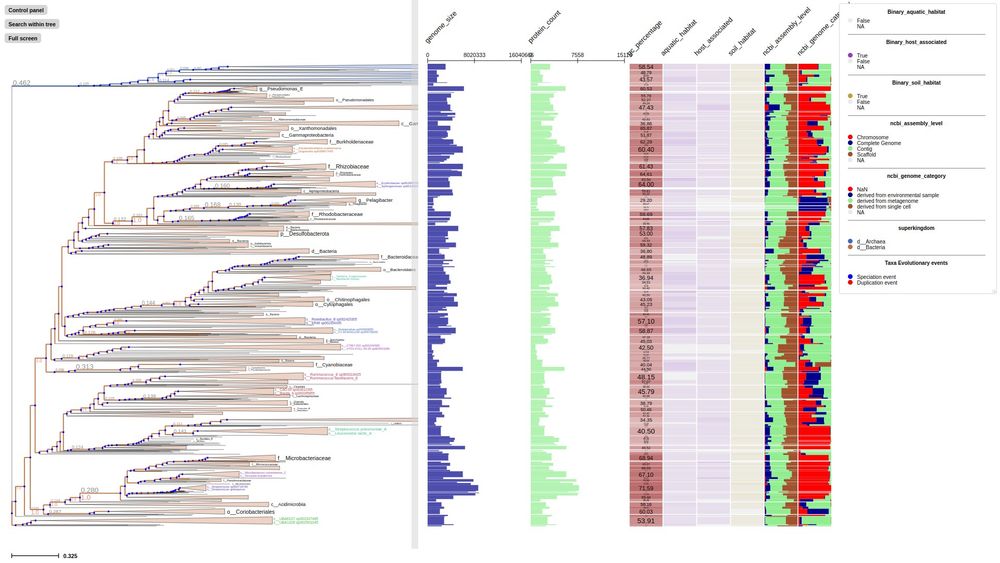
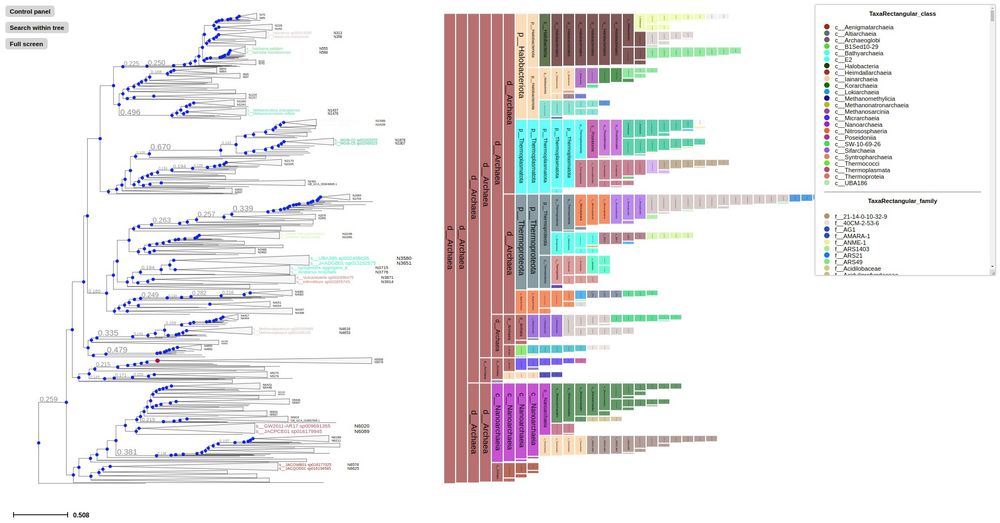
First time to cover new ETE4 features: Interactive viz of huge tree, profiling...
In Madrid, April 21-25. Travel fellowships available!
gut.bmj.com/content/...

gut.bmj.com/content/...

